Figure 4.
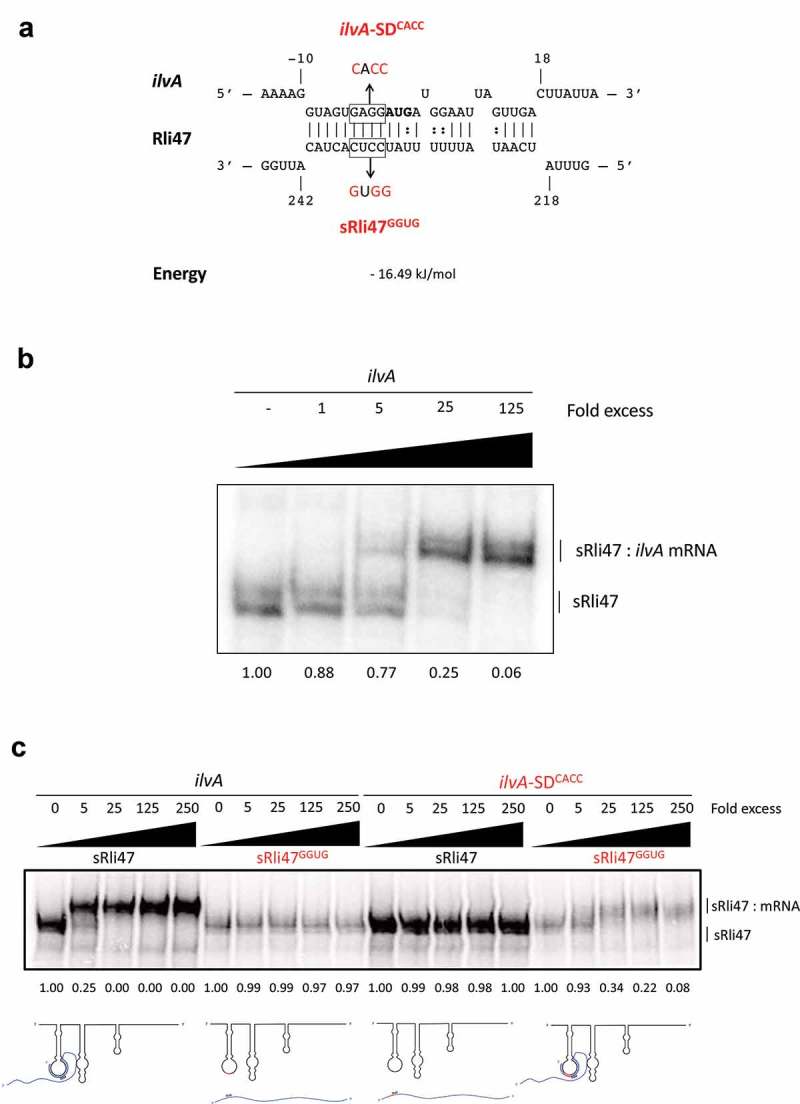
Mutational Analysis of truncated Rli47 and ilvA mRNA base pairing by Electrophoretic Mobility Shift Assay. (a) Predicted basepairing between the SD region of ilvA mRNA and Rli47. The mutated nucleotides are shown in red and the sequences of the minimal mutant variants ilvA-SDCACC and sRli47GGUG are indicated. (b) Electrophoretic mobility shift assays (EMSAs) of the interaction between ilvA mRNA and Rli47 truncated transcripts (sRli47). Truncated (sRli47) transcripts of Rli47 were incubated with increasing concentrations of unlabeled ilvA mRNA. (c) Secondary structure denaturation step was performed previous to binding incubation of labelled sRli47 and sRli47GGUG with increasing concentrations of unlabeled ilvA RNA or the mutant ilvA-SDCACC. Similar results were shown even without the denaturation of the secondary structures (Fig. S5). The fraction of unbound Rli47 is shown below each lane. AUG denotes the start codon of ilvA.
