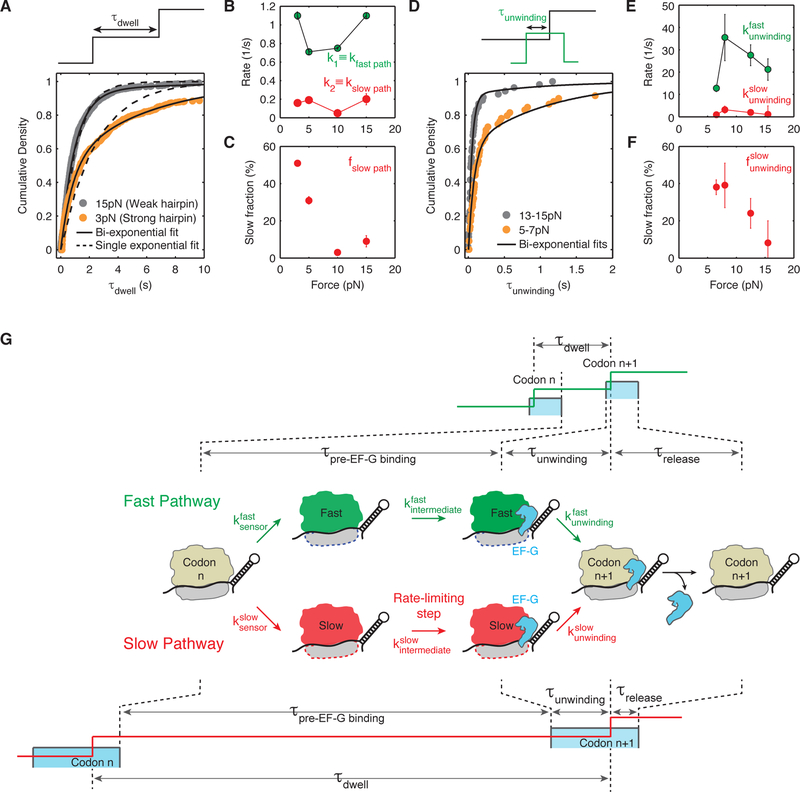
Figure 4. Ribosome translates through a hairpin via two parallel pathways that bifurcate before EF-G binding and converge after hairpin opening.
(A) Cumulative density of τdwell, the time ribosome spends at each codon, is best fit by a mixture of two exponentials given by: .
(B) Summary of fast and slow pathway rates, kfast path and kslow path obtained from fits to the cumulative distribution of τdwell at forces 3 pN (n = 551 events, 19 molecules), 5 pN (n = 256 events, 8 molecules), 10 pN (n = 776 events, 19 molecules) and 15 pN (n = 889 events, 24 molecules). Error bars represent 95% confidence intervals. Concentration of EF-G is 10 µM. kfast path and kslow path, remain constant at ~0.92 codon/s and ~0.15 codon/s respectively at various forces.
(C) Fraction of events going through the slow pathway, ƒslow path, as obtained from fitting shown in (A). ƒslow path increases from ~10 % at high force (> 10 pN) to ~30–50 % at low force (< 7 pN). Error bars represent 95% confidence intervals.
(D) Cumulative density of τunwinding, the time between EF-G binding and hairpin opening, is best fit by a mixture of two exponentials given by: .
(E) Summary of bi-exponential rates and obtained from fits to the cumulative distribution of τunwinding at forces 5–8 pN (n = 62 events, 14 molecules), 7–9 pN (n = 46 events, 10 molecules), 12–13 pN (n = 80 events, 20 molecules) and 14–17 pN (n = 53 events, 22 molecules). These experiments were performed in passive mode, i.e., the force is maintained in a particular regime rather than being held constant. Concentration of EF-G is 10 nM. Error bars represent 95% confidence intervals.
(F) Fraction of events going through the slow unwinding pathway, , increase from ~15% at high force (> 12 pN) to ~40% at low force (< 9 pN). Error bars represent 95% confidence intervals.
(G) Proposed kinetic scheme: first, the ribosome ‘senses’ the hairpin barrier and irreversibly switches into either a ‘fast’ state (shown in green) or a ‘slow’ state (shown in red) via rates or respectively. The ratio, , fraction of translation events that go through either pathway as shown in (C) and (F). Then, the ribosome in either ‘fast’ or ‘slow’ state must undergo an intermediate transition ( or ) that becomes rate limiting at low force and determines the overall rates shown in (B). It is possible that this intermediate transition is the rate of EF-G binding (see Discussion). Then, the ribosome unwinds the hairpin via rates and shown in (E). Finally, EF-G is released in a similar fashion in either pathway suggesting that the bifurcated pathways converged upon unwinding (Figure S6). A cartoon of the fleezers trajectory for both the fast and slow pathway is shown above and below the kinetic scheme. Notice that while τunwinding increases in the slow pathway, it does not increase enough to account for the total increase in τdwell.