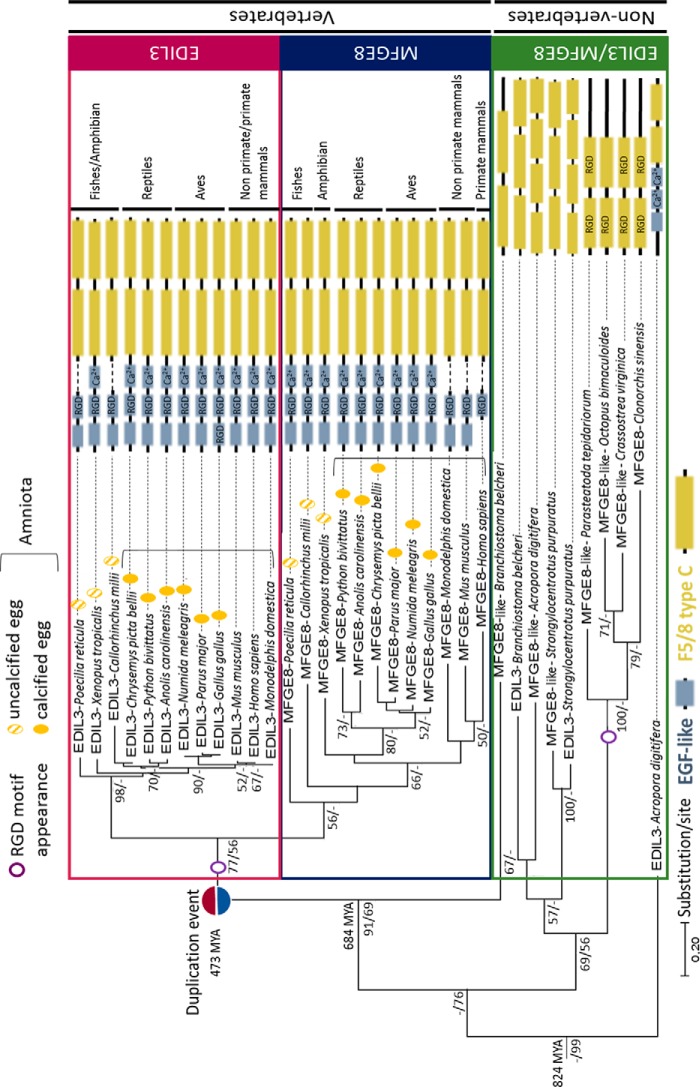
Figure 2.
Phylogenetic tree reconstruction of EDIL3 and MFGE8 common evolutionary histories in Eumetazoa. Multiple alignment of protein sequences (LG substitution model) was performed using Clustal Omega, and the phylogenetic tree was reconstructed using Maximum Likelihood (in MEGA7) and Bayesian inference (in BEAST version 1.10.4) methods. The topology of the tree is that resulting from Maximum Likelihood. Node values correspond to the percentage of Maximum Likelihood bootstrap values/Bayesian posterior probabilities, respectively. Only values above 50 are indicated. The divergence times were analyzed on Timetree. The schematic representation of EDIL3 and MFGE8 protein domains were acquired with HomoloGene of NCBI and several databases (ExPASy-PROSITE, Ensembl, and SMART). RGD, integrin-binding site; Ca2+, calcium-binding site. NCBI accession numbers for each protein are listed in Table S3.