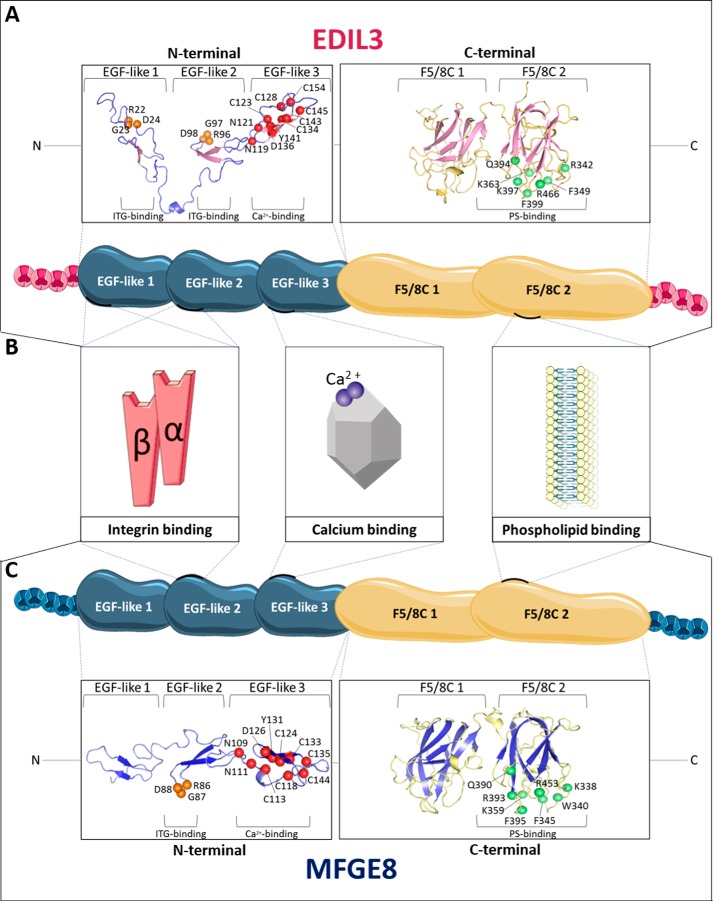
Figure 6.
In silico prediction and structure modeling of EDIL3 and MFGE8 protein domains and binding sites. A, tertiary structure prediction of EGF-like and F5/8C regions of G. gallus EDIL3 (XP_424906.3). B, schematic representation of G. gallus EDIL3 and MFGE8 protein domains, including their binding sites. C, tertiary structure prediction of EGF-like and F5/8C regions of G. gallus MFGE8 (NP_001264039.1). The tertiary structure prediction models were generated by EGF-like and F5/8C domain regions splitting using the I-TASSER template-based modeling. Following prediction modeling, the generated tertiary structures were edited using PyMOL. Orange balls indicate RGD residues of the ITG-binding sites; red balls illustrate residues of the Ca2+-binding sites, and green balls display PS-binding sites. The position of the residues is indicated according to their location in the entire protein. Elements were from Servier Medical Art, licensed under a Creative Commons Attribution 3.0 Unported License.