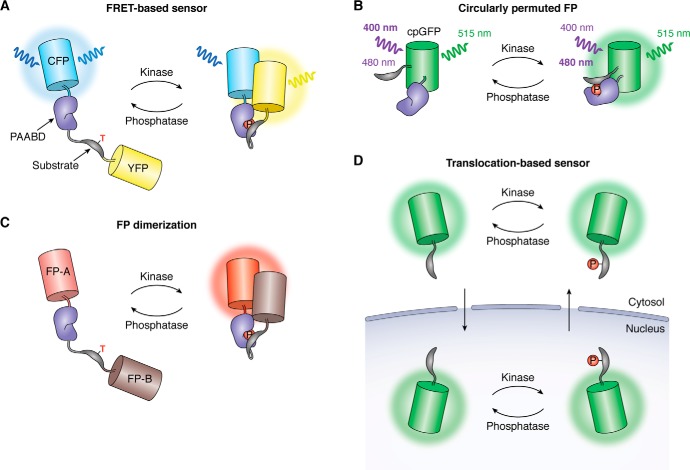
Figure 1.
Genetically encoded biosensors for visualizing kinase activity in living cells. A, FRET-based biosensors utilize a kinase-inducible molecular switch to modulate the relative distance and orientation of a pair of FPs in response to changes in biosensor phosphorylation, thereby coupling endogenous kinase activity to biosensor FRET efficiency. As shown here, the molecular switch is often constructed by fusing a substrate peptide for a specific kinase to a PAABD, such that recognition of the phosphorylated substrate by the PAABD drives a conformational change in the biosensor. B, single-fluorophore kinase biosensors have recently been generated in which the substrate peptide and PAABD are inserted within the sequence of a cpFP, whereby the recognition of the phosphorylated substrate by the PAABD distorts the barrel of the cpFP, thus altering its photophysical behavior. In the example shown here, phosphorylation of the sensor leads to a shift in the maximum excitation wavelength of cpGFP from 400 to 480 nm, producing an excitation-ratiometric response. C, an alternative strategy for constructing single-fluorophore biosensors retains the same overall configuration of FRET-based sensors but replaces the FRET FP pair with a pair of dimerization-dependent FPs. This design also relies on the molecular switch to modulate the proximity of the dimerization-dependent FPs, as bringing FP-A and B into close proximity results in enhanced fluorescence emission by FP-A and has been used to develop red-fluorescent kinase sensors. D, instead of relying on a conformational change, translocation-based kinase biosensors use phosphorylation to control the behavior of nuclear localization and export signals that are incorporated within the substrate sequence. At rest, the dephosphorylated biosensor accumulates in the nucleus, whereas elevations in kinase activity drive the translocation of the biosensor into the cytosol, whereby the ratio of nuclear-to-cytoplasmic fluorescence serves as a dynamic readout of kinase activity.