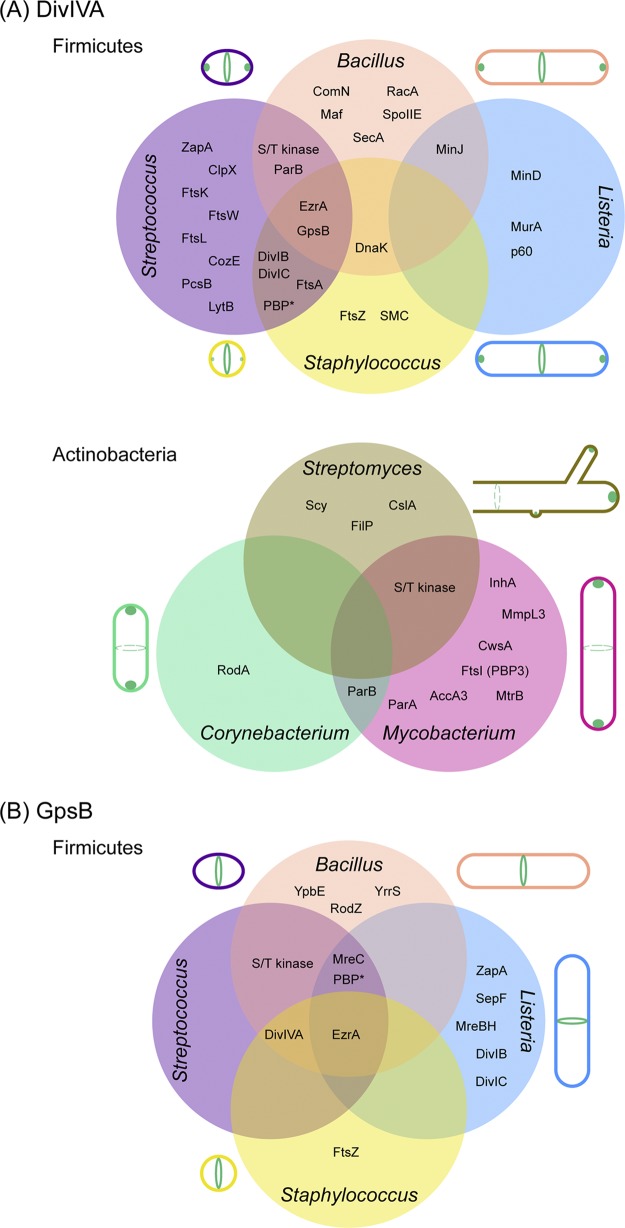
FIG 1.
Localization of DivIVA and GpsB and the overlapping network of their interaction partners in Gram-positive bacteria. Venn diagrams show the interaction partners of DivIVA (A) and GpsB (B) discussed in this article. (A) (Top panel) Clockwise from the top right, localization patterns of DivIVA in actively dividing cells of B. subtilis, L. monocytogenes, S. aureus, and S. pneumoniae are shown. (Bottom panel) Clockwise from the top, localization patterns of DivIVA in actively growing cells of Streptomyces, Mycobacterium, and Corynebacterium species are depicted. Dashed circles indicate alternate sites of localization. The Streptomyces cell is depicted in an open-ended manner to indicate the mycelial growth pattern. (B) Clockwise from the top right, localization patterns of GpsB in actively dividing cells of B. subtilis, L. monocytogenes, S. aureus, and S. pneumoniae are presented. The subcellular locations of DivIVA and GpsB are illustrated in green. A comprehensive list of DivIVA and GpsB partners and methods used to test interactions, including negative results, is provided in Table S1. *, the specific names of penicillin-binding proteins are shown in Table S1.