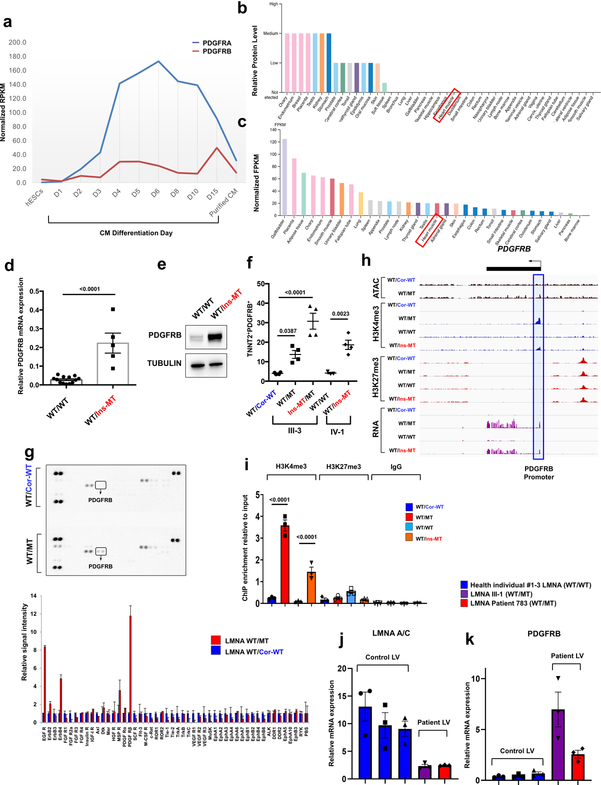
Extended Data Fig. 9. PDGFRB is up-regulated in LMNA-mutant iPSC-CMs.
a, Expression levels of PDGFRA and PDGFRB during human iPSC-CM differentiation process. The data were adapted from GSE76523. b, c, Protein and RNA levels of PDGFRB in human tissues. The data were adapted from Protein and RNA Atlas Database. d, Real-time PCR analysis of PDGFRB expression in LMNA-mutant and control iPSC-CMs. Data are expressed as mean ± s.e.m., and a two-tailed Student’s t-test was used to calculate P values. n=13 (WT/WT), n=5 (WT/Ins-MT). Numbers above the line show significant P values. e, Immunoblot analysis of PDGFRB protein levels in control versus mutant iPSC-CMs. GAPDH was used as loading control. The experiments were repeated twice independently with similar results. f, Flow cytometry analysis of TNNT2+ and PDGFRB+ cells in control and mutant iPSC-CMs. n=4. g, Kinase array of control and mutant iPSC-CMs. Fifty different protein kinases were presented in each chip. Raw images of blotting membrane (left). Two dots carried same antibody for technical duplicate. Quantification of signal intensity of each spot (right). h, Representative images of ChIP-seq, ATAC-seq, and RNA-seq on the genomic regions of PDGFRB. Blue box represents the promoter region of PDGFRB. i, ChIP-qPCR of H3K4me3 and H3K27me3 enrichment at the promoter region of PDGFRB in control and mutant iPSC-CMs. n=3. j, k, Real-time PCR analysis of LMNA and PDGFRB expression levels in left ventricle heart tissue from health controls (n=3) and LMNA-DCM patients (n=2). Data are expressed as mean ± s.e.m. The Kinase data in g was repeated twice independently with similar results. f, i, Data are expressed as mean ± s.e.m., and statistical significance was obtained using one-way ANOVA. Numbers above the line show significant P values.