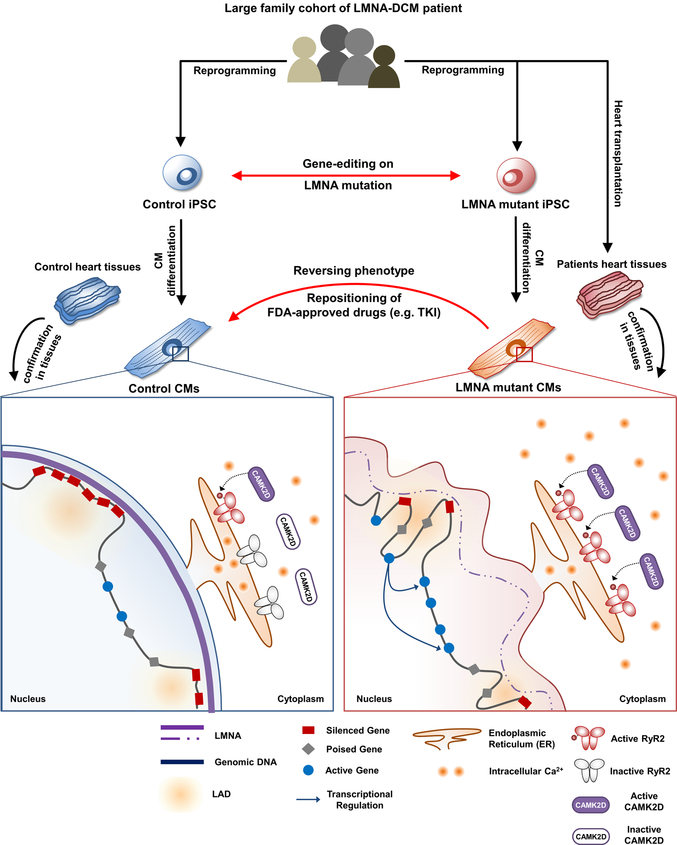
Extended Data Fig. 12. Proposed disease model of LMNA-DCM.
We recruited a large family cohort with DCM and generated patient-specific induced pluripotent stem cells (iPSCs) from several patients (n=5) and healthy individuals (n=2). We next utilized gene-edited isogenic iPSC lines (n=4) and patient heart tissues to address the intriguing question why patients with LMNA-DCM have increased manifestation of cardiac arrhythmias. The electrophysiological studies of mutant iPSC-derived cardiomyocytes (iPSC-CMs) demonstrated that LMNA mutation was the cause of increased arrhythmogenicity in LMNA-mutant iPSC-CMs. We also found that the LMNA mutation caused haploinsufficiency in LMNA, which led to abnormal calcium homeostasis in mutant iPSC-CMs through up-regulation of calcium handling genes. Whole transcriptome profiling (RNA-seq) further demonstrated an abnormal activation of PDGF pathway in mutant iPSC-CMs. The inhibition of PDGF signal pathway by treatment of siRNA or FDA-approved drugs such as sunitinib and crenolanib could rescue the arrhythmic phenotype of LMNA-mutant iPSC-CMs. Cross-analysis of various ChIP-seq, ATAC-seq, and RNA-seq revealed a possible underlying mechanism that haploinsufficiency of LMNA could disrupt global chromatin conformation, resulting in abnormal gene expression of mutant iPSC-CMs. These findings were further corroborated by studies in cardiac tissues from healthy and LMNA-DCM patients, thus validating a novel mechanism of LMNA-DCM pathogenesis both in vitro and in vivo.