Fig. 7.
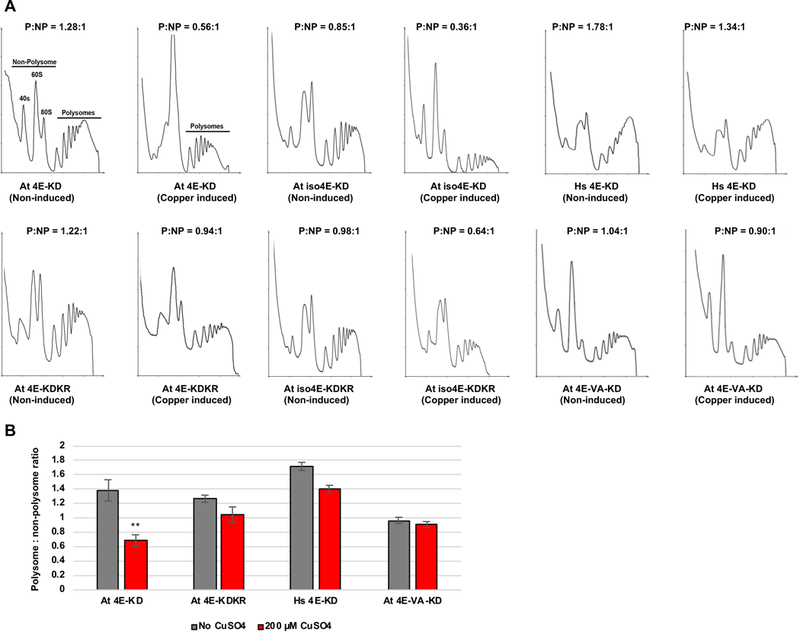
SnRK1-KD expression inhibits polysome formation in complemented yeast cells in a phosphorylation site-dependent manner. (A) The indicated Jo55 cdc33Δ-based yeast strains lacking endogenous eIF4E were incubated in the absence or presence of 200 μM CuSO4 to induce SnRK1-KD or SnRK1-KDKR (control) expression, and cell extracts were analyzed on sucrose density gradients to obtain polysome profiles. A polysome:non-polysome ratio (P:NP) was calculated for each sample. Representative traces are shown. Dramatic reductions in P:NP ratio were observed following SnRK1-KD induction in complemented strains At 4E-KD and At iso4E-KD, but not At 4E-VA-KD or Hs 4E-KD. (B) Histogram shows average P:NP ratios for At 4E-KD and At 4E-KDKR, Hs 4E-KD, and At4E-VA-KD obtained with and without copper induction. Error bars represent standard error of the mean. Asterisks indicate the difference between uninduced and copper induced At 4E-KD samples is significant at the p < 0.01 level, as determined by Student’s t-test.
