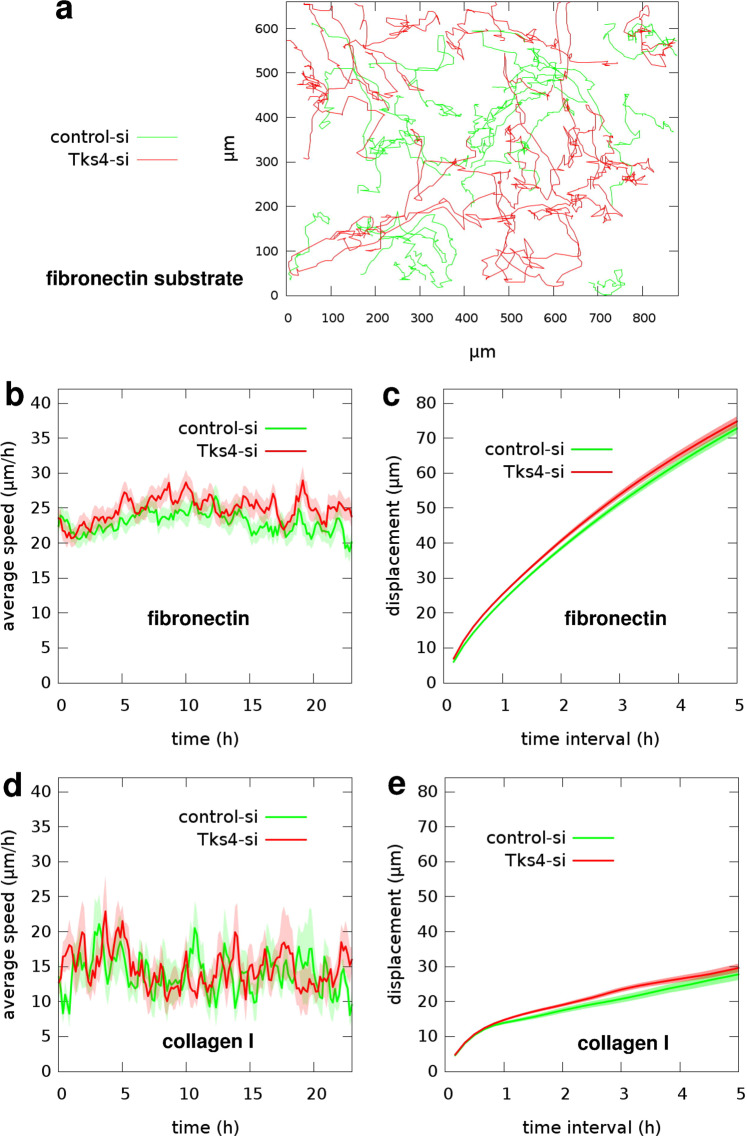
Figure 3.
Random motility of endothelial cells on 2D substrates. (a) Representative cell trajectories observed in two parallel HUVEC cultures – transfected with control-siRNA (green) or Tks4-siRNA (red) – on fibronectin substrate are superimposed for comparison. Trajectories were extracted from 24 hours long time-lapse recordings, also see Supplementary Videos S1 and S2. (b) Time-dependent cell speeds on fibronectin, averaged over the tracked cells for each time point. (c) Average cell displacements on fibronectin during various time intervals. Data in (b and c) were collected from n = 145 control-si cells and n = 151 Tks4-si cells on fibronectin in two independent sets of experiments. (d) Time-dependent cell speeds on collagen I. (e) Average cell displacements on collagen I. Data in (d and e) were collected from n = 20 control-si cells and n = 20 Tks4-si cells on collagen I. Error stripes in graphs correspond to SEM.