Abstract
Background: Obstructive sleep apnea (OSA) increases the risk of Alzheimer’s disease (AD), and inflammation may be involved in the early pathogenesis of AD in patients with OSA. However, the potential pathways between OSA and AD have yet to be established. In this study, we aimed to investigate differential expressions of AD-associated genes in OSA patients without evident AD or dementia. Methods: This prospective case-control study included five patients with severe OSA and five age and sex-matched patients with non-severe OSA without evident dementia who underwent uvulopalatopharyngoplasty between 1 January 2013 and 31 December 2015. The expressions of genes associated with AD were analyzed using whole-exome sequencing. Unsupervised two-dimensional hierarchical clustering was performed on these genes. Pearson’s correlation was used as the distance metric to simultaneously cluster subjects and genes. Results: The expressions of CCL2, IL6, CXCL8, HLA-A, and IL1RN in the patients with severe OSA were significantly different from those in the patients with non-severe OSA and contributed to changes in the immune response, cytokine–cytokine receptor interactions, and nucleotide-binding oligomerization domain-like receptor signaling pathways. Conclusions: Inflammation may contribute to the onset of AD and physicians need to be aware of the potential occurrence of AD in patients with severe OSA.
Keywords: Alzheimer’s disease, inflammation, obstructive sleep apnea, transcriptome
1. Introduction
Obstructive sleep apnea (OSA) can cause intermittent hypoxia, reoxygenation, hypercapnia, hypocapnia, changes in cerebral blood flow, and sleep fragmentation, which may result in brain disease [1]. The association between OSA and Alzheimer’s disease (AD) has been suggested based on population-based data and meta-analysis studies [2,3,4,5,6]. Furthermore, the potential pathways and causal relationships between OSA and AD have yet to be established [7].
In the last decade, “high-throughput” transcriptome investigation technologies have been developed to identify co-regulated gene networks linked with biological pathways and potentially modulating disease predisposition, outcomes, and progression [8]. For example, early-onset AD (EOAD), constituting barely 1–2% of all cases of AD and being diagnosed before the age of 65 years, is usually autosomal dominant inherited with genes including APP [9], FKBP1B [10], PSEN1 [11,12] and PSEN2 [11,12] being regarded as major factors. In comparison, the association between late-onset AD (LOAD) and genetic variants is much more complex, with at least 56 LOAD-associated genes [9,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36] reported in the literature. These genes may involve lipid metabolism and regulate the production and clearance of amyloid β, thereby inducing AD [37]. Previous studies also showed an association of OSA and AD-related biomarkers, including brain amyloid β load and cerebrospinal fluid levels of tau [38,39,40,41,42].
In this study, we aimed to investigate differential expressions of genes associated with AD using whole-exome sequencing in middle-aged patients with OSA without evident AD using a case-control study.
2. Methods
2.1. Ethical Considerations
This study was approved by the Institutional Review Board of the Chang Gung Memorial Foundation (101-3547A3 and 102-5609A3), and written informed consent was obtained from all participants.
2.2. Participants
We prospectively recruited 10 consecutive otherwise healthy patients without memory deficits (five severe OSA cases with an apnea–hypopnea index [AHI] ≥30 events/h and five non-severe OSA controls with an AHI <15 events/h) who underwent uvulopalatopharyngoplasty in a tertiary care hospital between 1 January 2013 and 31 December 2015 [43]. None of the patients had a Mini-Mental State Examination score <25 [44], stroke, or heart failure after 2 years of follow-up. Resected submucosal tissues were obtained intraoperatively from the uvula.
2.3. RNA Isolation of the Study
Samples were immediately rinsed with phosphate-buffered saline to remove excess blood, minced, subjected to RNA extraction, and stored at −80 °C. We isolated and sequenced the total RNA of resected submucosal tissues of the uvula using RNA-Seq technology [45]. In brief, total RNA from cells of identical density was isolated using guanidinium phenol reagent (TRIzol reagent; Life Technologies, Grand Island, NY, USA) according to the manufacturer’s instructions. After synthesis, purification, and 3’-end single nucleotide A (adenine) addition of double-strand cDNA, the fragments were enriched by PCR amplification. During the quality control step, an Agilent 2100 Bioanaylzer (Agilent Technologies, Santa Clara, CA) and ABI StepOnePlusTM Real-Time PCR System (Applied Biosystems, Foster City, CA) were used to qualify and quantify the sample library. The library products were ready for sequencing via an Illumina HiSeqTM 2000. On average, 22,415,673 raw sequencing reads were generated and then 22,181,205 clean reads were obtained after filtering those of low quality. After obtaining raw data, quality control of alignment was performed to determine whether resequencing was needed.
2.4. Bioinformatics Analysis
After passing quality control, the alignment data were used to calculate the distribution of reads on reference genes and mapping ratio. We then performed downstream analysis including gene expression and deep analysis based on the gene expression. Gene quantification was performed using RNA-Seq by Expectation Maximization [46] to determine which transcripts were isoforms of the same gene, and gene expression levels were calculated using the method of fragments per kilobase of transcript per million map reads for direct comparisons of differences in gene expressions among samples. Gene expression values across all subjects were normalized as follows: Gnorm = (expression level of the gene for each subject – average expression level of the gene across all subjects)/standard deviation.
Differentially expressed genes (DEGs) were screened using the Poisson distribution method [47], and correction for type I and II errors was performed using the false discovery rate (FDR) method [48]. The Q value (error ratio) was calculated as the number of false positive genes/the number of selected DEGs. More stringent criteria (FDR ≤ 0.001 and an absolute value of Log2Ratio ≥ 1) were applied to identify DEGs. For our analysis, 63 genes (for details, see Table S1 in the Supplementary Materials) including four EOAD-associated genes (APP [9], FKBP1B [10], PSEN1 [11,12], and PSEN2 [11,12]) and 56 LOAD-associated genes (ABCA1 [13,14], ABCA7 [13,14], ANK1 [15], APOC1 [16], APOE [17], BACE2 [18], BCR [16], BIN1 [19], CASS4 [20], CCL2 (also known as monocyte chemoattractant protein 1 [MCP-1]) [21], CD2AP [13,22], CD33 [13,22], CELF1 [20], CLU [23,24], COX4I1 [25], CR1 [24], CXCL8 [35], DSG2 [20], EPHA1 [13,22,26], EPHA10 [13,22,26], FAM136A [16], FERMT2 [20], GAS7 [27], GRIK2 [16], GRN [28], HAS3 [16], HLA-A [36], HLA-DRB1 [20], HLA-DRB5 [20], IL1RN [34], IL6 [29], INPP5D [20], ITGAL [16], KLK2 [16], LRP2 [30], MAPT [28], MEF2C [20], MS4A4 [13,19,22], MS4A6E [13,19,22], PICALM [23], PILRA [16], PIN1 [32], PLD3 [9], PRSS42 [16], PRSS45 [16], PTK2B [20], RIN3 [20], SEC31A [16], SLC22A2 [16], SLC24A4 [20], SORL1 [20], STOX2 [16], THNSL2 [16], TMIE [16], TRDMT1 [16], TREM2 [33] and ZCWPW1 [20]) were selected from well-established curated resources, including published literatures, the Database for Annotation, Visualization and Integrated Discovery (DAVID) (version 6.8; https://david.ncifcrf.gov/) [49], Gene Association Database (GAD) (https://geneticassociationdb.nih.gov/), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) [50]. We performed a deep analysis based on DEGs, including GAD, Gene Ontology (GO) enrichment analysis, and KEGG pathway enrichment analysis.
2.5. Statistical Analysis
Unsupervised two-dimensional hierarchical clustering was performed on these 63 unique genes. Pearson’s correlation was used as the distance metric to simultaneously cluster subjects (based on their global expression profile) and genes (based on expression levels across subjects). The calculated p-values of bioinformatics analysis were subjected to Benjamini correction, taking an adjusted p-value < 0.05 as a threshold. All analyses were performed using SAS software version 9.4 (SAS Institute, Cary, NC, USA) and SPSS software version 24 (International Business Machines Corp., Armonk, NY, USA), and the level of statistical significance was set at p < 0.05.
3. Results
3.1. Participant Characteristics
The patient characteristics are shown in Table 1. The patients were relatively younger, mostly male and non-obese, but there were no significant differences between the two groups in age, male sex, body mass index, minimal SpO2, or time with SpO2 < 85%. As expected, the patients with severe OSA had a significantly higher mean AHI (mean, 60.6; SD, 21.2) compared to those with non-severe OSA (mean, 7.0; SD, 4.3) (effect size = 3.50). None of the participants had a diagnosis of the comorbidities associated with AD.
Table 1.
Differences in patient characteristics between the severe obstructive sleep apnea (OSA) and non-severe OSA groups.
| Variables | Severe OSA | Non-Severe OSA | Effect Size | p-Value |
|---|---|---|---|---|
| Patients | n = 5 | n = 5 | ||
| Age (years), mean (SD) | 34.6 (4.2) | 41.2 (12.0) | 0.73 | 0.07 |
| Male sex, n (%) | 5 (100) | 4 (80) | >0.99 | |
| BMI (kg/m2), mean (SD) | 26.4 (3.2) | 25.5 (2.9) | 0.30 | 0.07 |
| AHI (events/h), mean (SD) | 60.6 (21.2) | 7.0 (4.3) | 3.50 | <0.001 |
| Minimum SpO2 (%), mean (SD) | 79.0 (6.0) | 85.2 (2.3) | 1.37 | 0.06 |
| Time SpO2 < 85% (%), mean (SD) | 1.66 (2.3) | 0.04 (0.04) | 1.00 | 0.15 |
Abbreviations: AHI: apnea–hypopnea index; BMI: body mass index; OSA, obstructive sleep apnea; SpO2: oxygen saturation measured by pulse oximetry.
3.2. AD-Associated Transcriptomic Expression in the Uvular Tissue of the Patients with Severe OSA
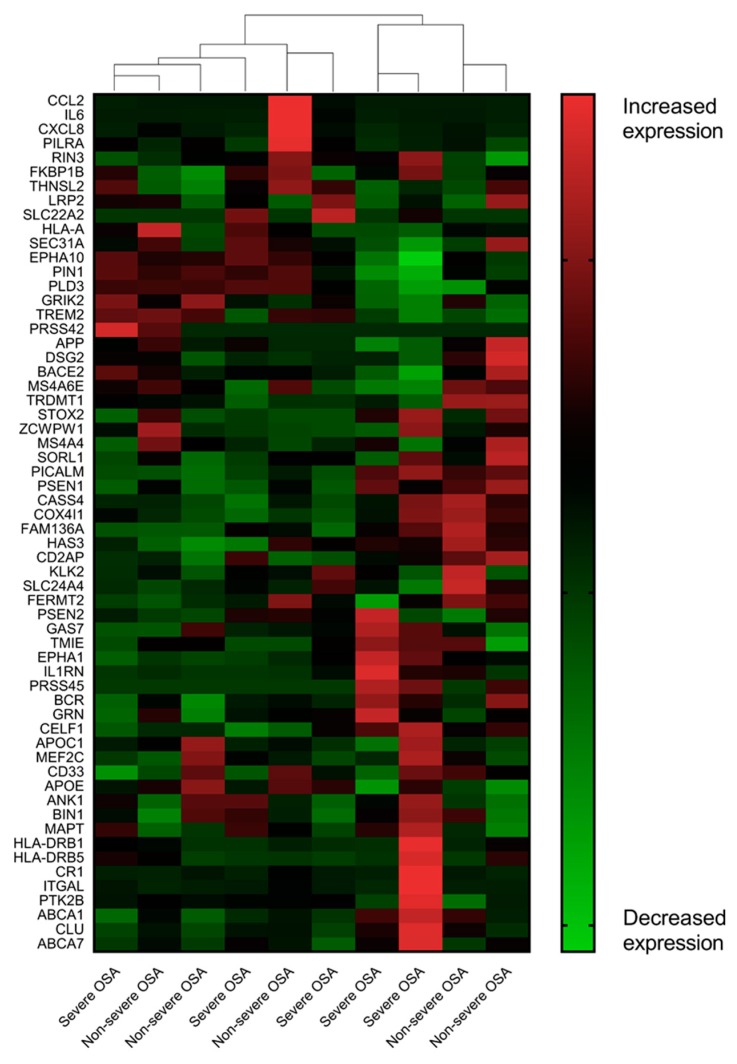
Unsupervised two-dimensional hierarchical clustering of the expression profiles from over 60 AD-associated genes did not differentiate the patients with severe OSA from those with non-severe OSA (Figure 1). We identified five AD-associated DEGs including one up-regulated gene (IL1RN) and four down-regulated genes (CCL2, CXCL8, HLA-A, and IL6) at a FDR of 0.001 (Table 2). None of the EOAD-associated genes were significantly differently expressed. Detailed comparisons of the expressions of the 60 AD-associated genes between the two groups in descending order by probability are shown in Table S2 in the Supplementary Materials.
Figure 1.
Alzheimer’s disease-associated transcriptome of the uvula tissue from the patients with severe OSA and non-severe OSA. In this analysis, we made no a priori designation of a subject’s phenotype (severe OSA, non-severe OSA control) when clustering individuals based on their oropharyngeal gene expression patterns. The results implied that severe sleep apnea elicited modest perturbations in the AD-associated transcriptional response of the oropharynx.
Table 2.
Expression of representative Alzheimer’s disease-associated genes among the patients with severe OSA and non-severe OSA in descending order by probability.
| Gene Symbol | Expression | Log2(Fold Change) | Regulation | Probability | |
|---|---|---|---|---|---|
| Severe OSA | Non-Severe OSA | ||||
| Early-onset Alzheimer’s disease | |||||
| APP | 382.418 | 587.644 | −0.619 | Down | 0.503 |
| PSEN1 | 12.776 | 14.876 | −0.220 | Down | 0.286 |
| PSEN2 | 7.204 | 6.208 | 0.215 | Up | 0.265 |
| FKBP1B | 3.786 | 4.126 | −0.124 | Down | 0.216 |
| Late-onset Alzheimer’s disease | |||||
| CCL2 | 12.992 | 116.716 | −3.167 | Down | 0.868 a |
| IL6 | 2.854 | 33.06 | −3.534 | Down | 0.857 a |
| CXCL8 | 2.042 | 20.058 | −3.296 | Down | 0.824 a |
| HLA-A | 1.272 | 14.006 | −3.460 | Down | 0.811 a |
| IL1RN | 239.35 | 57.236 | 2.064 | Up | 0.806 a |
| CLU | 490.412 | 308.192 | 0.670 | Up | 0.522 |
| BIN1 | 23.218 | 18.884 | 0.298 | Up | 0.335 |
| APOE | 24.526 | 29.308 | −0.257 | Down | 0.318 |
| SORL1 | 8.136 | 9.338 | −0.199 | Down | 0.267 |
| PICALM | 56.394 | 52.512 | 0.103 | Up | 0.233 |
Abbreviations: OSA, obstructive sleep apnea. a Significantly up- and down-regulated genes were defined as false discovery rate (FDR) ≤ 0.001 (i.e., probability ≥ 0.8) and an absolute value of Log2Ratio ≥ 1.
Highly enriched modules, relating to these five DEGs in the patients with severe OSA, included 27 biological processes, three cellular components, and three molecular functions (for details, see Table S3 in the Supplementary Materials). Furthermore, only six biological processes (immune response; cellular response to interleukin-1; cellular response to tumor necrosis factor; cellular response to lipopolysaccharide; regulation of vascular endothelial growth factor production; and negative regulation of IL-1-mediated signaling pathway) and one cellular component (extracellular space) reached statistical significance after Benjamini correction.
We found a highly enriched AD module as derived from “GAD-disease” in the patients with severe OSA (five counts; DEGs with pathway annotation: 1.3%; unadjusted p-value = 4.3 × 10−7; Benjamini adjusted p-value = 4.6 × 10−4). However, functional annotation as derived from, the KEGG, did not reveal an enriched AD module (Table 3). Nevertheless, the nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway and cytokine–cytokine receptor interaction pathway were significantly enriched.
Table 3.
Functional annotation chart of the Kyoto Encyclopedia of Genes and Genomes according to the five Alzheimer’s disease-associated differentially expressed genes in severe OSA patients.
| Term | Count | Percentage | p-Value | Adjusted p-Value |
|---|---|---|---|---|
| Malaria | 3 | 0.8 | <0.001 | 0.007 a |
| NOD-like receptor signaling pathway | 3 | 0.8 | <0.001 | 0.005 a |
| Rheumatoid arthritis | 3 | 0.8 | <0.001 | 0.008 a |
| Chagas disease | 3 | 0.8 | <0.001 | 0.008 a |
| Influenza A | 3 | 0.8 | 0.002 | 0.02 a |
| Herpes simplex infection | 3 | 0.8 | 0.002 | 0.02 a |
| Cytokine–cytokine receptor interaction | 3 | 0.8 | 0.003 | 0.02 a |
| Graft-versus-host disease | 2 | 0.5 | 0.01 | 0.09 |
| Legionellosis | 2 | 0.5 | 0.02 | 0.12 |
| Pertussis | 2 | 0.5 | 0.03 | 0.15 |
| Salmonella infection | 2 | 0.5 | 0.04 | 0.15 |
| Amoebiasis | 2 | 0.5 | 0.045 | 0.18 |
| TNF signaling pathway | 2 | 0.5 | 0.045 | 0.18 |
| Toll-like receptor signaling pathway | 2 | 0.5 | 0.045 | 0.18 |
| Hepatitis B | 2 | 0.5 | 0.06 | 0.22 |
| Non-alcoholic fatty liver disease | 2 | 0.5 | 0.06 | 0.21 |
| Transcriptional misregulation in cancer | 2 | 0.5 | 0.07 | 0.22 |
| Chemokine signaling pathway | 2 | 0.5 | 0.08 | 0.23 |
Abbreviations: NOD, nucleotide-binding oligomerization domain; OSA, obstructive sleep apnea; TNF, tumor necrosis factor; a p-values < 0.05 adjusted using the Benjamini method were statistically significant.
4. Discussion
In this study, five AD-associated DEGs were identified in our middle-aged patients with severe OSA without symptoms of AD. These findings suggest that preclinical changes in genetic expressions may contribute to the onset of AD in patients with severe OSA. The uvular tissue of the patients with severe OSA exhibited enriched biological processes that resulted in the change in state or activity of a cell in terms of movement, secretion, enzyme production, and gene expression because of an IL-1, tumor necrosis factor, or lipopolysaccharide stimulus. In the study, major genetic factors for EOAD (such as APP, FKBP1B, PSEN1, and PSEN2) and LOPD (such as APOE, CLU, SORL1, PICALM, and BIN1 [9,10,11,12,51]) did not show significantly different expressions in the patients with severe OSA without evident AD. However, the expressions of CCL2, IL6, CXCL8, HLA-A, and IL1RN were significantly different in the patients with severe OSA compared to those with non-severe OSA.
Previous studies [21,29,36,52] have reported significant associations between CCL2, HLA-A2, IL6, and IL1RN gene polymorphisms and AD, and IL6 gene polymorphisms have been associated with the risk of adult OSA [53]. We previously found that the expression of the CCL2 gene was significantly elevated in peripheral blood monocytes of patients with severe OSA [54], whereas its expression was significantly decreased in the uvular tissue in this study. Elevated serum levels of MCP-1, IL-1ß, and IL-6 in patients with OSA may also be risk factors for the development of AD [55,56,57]. The down-regulated expression of CXCL8 in patients with OSA has been reported to possibly reduce its protective effect against amyloid-ß-induced neurotoxicity [35]. Moreover, changes in the cytokine–cytokine receptor interaction pathway [58] or NOD-like receptor signaling pathway [59] may activate the NOD-like receptor 3 inflammasome and promote neuroinflammation and brain pathologies [59]. These observations suggest that inflammation links OSA and AD.
Although the prevalence of OSA in patients with dementia has been estimated to be high, little is known about the association between OSA and AD with regards to disease severity [60]. In a comparative study of nursing home patients, the severe OSA group had significantly lower dementia rating scales than the mild–moderate OSA group (31 vs. 127, p < 0.004) [61]. The severe OSA group (AHI ≥ 40 events/h) had a higher incidence of AD than those with less severe OSA (AHI < 40 events/h). Ancoli-Israel et al. also reported that the respiratory disturbance index was significantly correlated with total dementia rating scale score (r = −0.37, p < 0.001) [61]. In their study, the correlation could be influenced by different reliability in night-to-night respiratory disturbance index (r = 0.68) and dementia rating scale score (r = 0.97). After correction for attenuation, the correlation coefficient between respiratory disturbance index and dementia rating scale score was estimated to be approximately −0.46. These data suggest that severe OSA patients have a higher incidence of AD and lower cognitive function, and that there is a moderate association in disease severity between OSA and AD.
There are limitations to this study. First, although the use of molecular expression from excised uvular tissue may be a surrogate to correlate potential pathways of OSA-related AD, further research to compare the molecular expression in uvular tissue in AD patients is warranted to confirm our results. Second, this is a case-controlled study; therefore, we could not follow up these patients and investigate the occurrence of AD years later. Third, the sample size was small. However, this study proved the alterations in AD-associated gene expression in severe OSA patients. Based on our findings, inflammatory processes related to severe OSA may be possible mechanisms to prevent AD. Prospective clinical trials are necessary to elucidate the causal relationship between OSA and AD and, moreover, to clarify whether continuous positive airway pressure (CPAP) therapy for OSA affects the prevalence and onset of AD.
5. Conclusions
In our middle-aged patients with severe OSA without evident AD, the genetic alterations in the biological processes and pathways suggest that inflammation is a possible early change leading to the occurrence of AD. Our study extends the OSA disease spectrum and should serve to remind physicians to be aware of the potential occurrence of AD in patients with OSA. Further research is warranted to explore the genetic characteristics of OSA-AD and investigate the effect of CPAP therapy on the onset and occurrence of AD.
Acknowledgments
The authors would like to thank the Wallace Academic Editing for editing this manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/8/9/1361/s1, Table S1: Genes and pseudogenes associated with Alzheimer’s disease. Table S2: Summary of the expressions of 60 Alzheimer’s disease-associated genes and pseudogenes in the severe OSA and non-severe OSA groups in descending order by probability. Table S3: Results of Gene Ontology related to five Alzheimer’s disease-associated differentially expressed genes in severe OSA patients.
Author Contributions
H.-Y.L., M.-S.T. and L.-A.L. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis; Concept and design: H.-Y.L., M.-S.T., C.-G.H., R.Y.L.W. and L.-A.L.; Acquisition, analysis, or interpretation of data: H.-Y.L., M.-S.T., C.-G.H., R.Y.L.W., L.-P.C. and L.-A.L.; Drafting of the manuscript: H.-Y.L., M.-S.T. and L.-A.L.; Critical revision of the manuscript for important intellectual content: H.-Y.L., M.-S.T., L.-A.L., C.-G.H., R.Y.L.W., L.-P.C., N.-H.C., C.-H.L., C.-H.L., C.-M.H., W.-N.C. and L.-A.L.; Statistical analysis: M.-S.T. and L.-A.L.; Obtained funding: H.-Y.L., M.-S.T. and L.-A.L.; Administrative, technical, or material support: H.-Y.L., C.-G.H., R.Y.L.W., L.-P.C. and C.-M.H.; Supervision: H.-Y.L., L.-P.C., and C.-M.H.
Funding
The study was financially supported by grants from the Ministry of Science and Technology, Taiwan, R.O.C. (NSC102-2314-B-182A-082) and Chang Gung Memorial Hospital, Taoyuan, Taiwan, R.O.C. (CMRPG3F1091 and CFRPG6J0051). The Health Information and Epidemiology Laboratory of Chiayi Chang Gung Memorial Hospital provided statistical assistance and support from the Maintenance project of the Center for Big Data Analytics and Statistics (grant CLRPG6G0041). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Conflicts of Interest
The authors have no other funding, financial relationships, or conflicts of interest to disclose.
References
- 1.Rosenzweig I., Glasser M., Polsek D., Leschziner G.D., Williams S.C.R., Morrell M.J. Sleep apnoea and the brain: A complex relationship. Lancet Respir. Med. 2015;3:404–414. doi: 10.1016/S2213-2600(15)00090-9. [DOI] [PubMed] [Google Scholar]
- 2.Pan W., Kastin A.J. Can sleep apnea cause Alzheimer’s disease? Neurosci. Biobehav. Rev. 2014;47:656–669. doi: 10.1016/j.neubiorev.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 3.Chang W.P., Liu M.E., Chang W.C., Yang A.C., Ku Y.C., Pai J.T., Huang H.L., Tsai S.J. Sleep apnea and the risk of dementia: A population-based 5-year follow-up study in Taiwan. PLoS ONE. 2013;8:e78655. doi: 10.1371/journal.pone.0078655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J.E., Yang S.W., Ju Y.J., Ki S.K., Chun K.H. Sleep-disordered breathing and Alzheimer’s disease: A nationwide cohort study. Psychiatry Res. 2019;273:624–630. doi: 10.1016/j.psychres.2019.01.086. [DOI] [PubMed] [Google Scholar]
- 5.Bubu O.M., Brannick M., Mortimer J., Umasabor-Bubu O., Sebastiao Y.V., Wen Y., Schwartz S., Borenstein A.R., Wu Y., Morgan D., et al. Sleep, Cognitive impairment, and Alzheimer’s disease: A Systematic Review and Meta-Analysis. Sleep. 2017:40. doi: 10.1093/sleep/zsw032. [DOI] [PubMed] [Google Scholar]
- 6.Shi L., Chen S.-J., Ma M.-Y., Bao Y.-P., Han Y., Wang Y.-M., Shi J., Vitiello M.V., Lu L. Sleep disturbances increase the risk of dementia: A systematic review and meta-analysis. Sleep Med. Rev. 2018;40:4–16. doi: 10.1016/j.smrv.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Macedo A.C., Balouch S., Tabet N. Is Sleep Disruption a Risk Factor for Alzheimer’s Disease? J. Alzheimer’s Dis. 2017;58:993–1002. doi: 10.3233/JAD-161287. [DOI] [PubMed] [Google Scholar]
- 8.Guffanti A., Simchovitz A., Soreq H. Emerging bioinformatics approaches for analysis of NGS-derived coding and non-coding RNAs in neurodegenerative diseases. Front. Cell. Neurosci. 2014;8:89. doi: 10.3389/fncel.2014.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruchaga C., Karch C.M., Jin S.C., Benitez B.A., Cai Y., Guerreiro R., Harari O., Norton J., Budde J., Bertelsen S., et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gant J.C., Blalock E.M., Chen K.C., Kadish I., Porter N.M., Norris C.M., Thibault O., Landfield P.W. FK506-binding protein 1b/12.6: A key to aging-related hippocampal Ca2+ dysregulation? Eur. J. Pharmacol. 2014;739:74–82. doi: 10.1016/j.ejphar.2013.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Q., Xie N., Tang B., Li R., Shen Y. Alzheimer’s Disease: From Genetic Variants to the Distinct Pathological Mechanisms. Front. Mol. Neurosci. 2017;10:319. doi: 10.3389/fnmol.2017.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alzheimer’s Association 2015 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2015;11:332–384. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina V., et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wavrant-De Vrieze F., Compton D., Womick M., Arepalli S., Adighibe O., Li L., Perez-Tur J., Hardy J. ABCA1 polymorphisms and Alzheimer’s disease. Neurosci. Lett. 2007;416:180–183. doi: 10.1016/j.neulet.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi S., Song J.H., Tan M.S., Zhang W., Wang Z.X., Jiang T., Tan L., Yu J.T. Association of Single-Nucleotide Polymorphism in ANK1 with Late-Onset Alzheimer’s Disease in Han Chinese. Mol. Neurobiol. 2016;53:6476–6481. doi: 10.1007/s12035-015-9547-x. [DOI] [PubMed] [Google Scholar]
- 16.Patel T., Brookes K.J., Turton J., Chaudhury S., Guetta-Baranes T., Guerreiro R., Bras J., Hernandez D., Singleton A., Francis P.T., et al. Whole-exome sequencing of the BDR cohort: evidence to support the role of the PILRA gene in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2017 doi: 10.1111/nan.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C.C., Liu C.C., Kanekiyo T., Xu H., Bu G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu M., Liu Y., Shen J., Lv D., Zhang J. Meta-analysis of BACE1 gene rs638405 polymorphism and the risk of Alzheimer’s disease in Caucasion and Asian population. Neurosci. Lett. 2016;616:189–196. doi: 10.1016/j.neulet.2016.01.059. [DOI] [PubMed] [Google Scholar]
- 19.Seshadri S., Fitzpatrick A.L., Ikram M.A., DeStefano A.L., Gudnason V., Boada M., Bis J.C., Smith A.V., Carassquillo M.M., Lambert J.C., et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flex A., Giovannini S., Biscetti F., Liperoti R., Spalletta G., Straface G., Landi F., Angelini F., Caltagirone C., Ghirlanda G., et al. Effect of proinflammatory gene polymorphisms on the risk of Alzheimer’s disease. Neurodegener. Dis. 2014;13:230–236. doi: 10.1159/000353395. [DOI] [PubMed] [Google Scholar]
- 22.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M.J., Tavernier B., et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 25.Lu J., Wang K., Rodova M., Esteves R., Berry D., Lezi E., Crafter A., Barrett M., Cardoso S.M., Onyango I., et al. Polymorphic variation in cytochrome oxidase subunit genes. J. Alzheimer’s Dis. 2010;21:141–154. doi: 10.3233/JAD-2010-100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lalli M.A., Garcia G., Madrigal L., Arcos-Burgos M., Arcila M.L., Kosik K.S., Lopera F. Exploratory data from complete genomes of familial alzheimer disease age-at-onset outliers. Hum. Mutat. 2012;33:1630–1634. doi: 10.1002/humu.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivasan K., Friedman B.A., Larson J.L., Lauffer B.E., Goldstein L.D., Appling L.L., Borneo J., Poon C., Ho T., Cai F., et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat. Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S.C., Pastor P., Cooper B., Cervantes S., Benitez B.A., Razquin C., Goate A., Ibero-American Alzheimer Disease Genetics Group Researchers. Cruchaga C. Pooled-DNA sequencing identifies novel causative variants in PSEN1, GRN and MAPT in a clinical early-onset and familial Alzheimer’s disease Ibero-American cohort. Alzheimer’s Res. 2012;4:34. doi: 10.1186/alzrt137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen L., Delabio R., Horiguchi L., Mizumoto I., Terazaki C.R., Mazzotti D., Bertolucci P.H., Pinhel M.A., Souza D., Krieger H., et al. Association between interleukin 6 gene haplotype and Alzheimer’s disease: A Brazilian case-control study. J Alzheimer’s Dis. 2013;36:733–738. doi: 10.3233/JAD-122407. [DOI] [PubMed] [Google Scholar]
- 30.Wang L.L., Pan X.L., Wang Y., Tang H.D., Deng Y.L., Ren R.J., Xu W., Ma J.F., Wang G., Chen S.D. A single nucleotide polymorphism in LRP2 is associated with susceptibility to Alzheimer’s disease in the Chinese population. Clin. Chim. Acta. 2011;412:268–270. doi: 10.1016/j.cca.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Chandrasekaran K., Giordano T., Brady D.R., Stoll J., Martin L.J., Rapoport S.I. Impairment in mitochondrial cytochrome oxidase gene expression in Alzheimer disease. Mol. Brain Res. 1994;24:336–340. doi: 10.1016/0169-328X(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 32.Arosio B., Bulbarelli A., Bastias Candia S., Lonati E., Mastronardi L., Romualdi P., Candeletti S., Gussago C., Galimberti D., Scarpini E., et al. Pin1 contribution to Alzheimer’s disease: Transcriptional and epigenetic mechanisms in patients with late-onset Alzheimer’s disease. Neurodegener. Dis. 2012;10:207–211. doi: 10.1159/000333799. [DOI] [PubMed] [Google Scholar]
- 33.Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S., Younkin S., et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarkowski E., Liljeroth A.-M., Nilsson Å., Minthon L., Blennow K. Decreased Levels of Intrathecal Interleukin 1 Receptor Antagonist in Alzheimer’s Disease. Dement. Geriatr. Cogn. Disord. 2001;12:314–317. doi: 10.1159/000051276. [DOI] [PubMed] [Google Scholar]
- 35.Ashutosh, Kou W., Cotter R., Borgmann K., Wu L., Persidsky R., Sakhuja N., Ghorpade A. CXCL8 protects human neurons from amyloid-beta-induced neurotoxicity: Relevance to Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2011;412:565–571. doi: 10.1016/j.bbrc.2011.07.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Listi F., Candore G., Balistreri C.R., Grimaldi M.P., Orlando V., Vasto S., Colonna-Romano G., Lio D., Licastro F., Franceschi C., et al. Association between the HLA-A2 allele and Alzheimer disease. Rejuvenation Res. 2006;9:99–101. doi: 10.1089/rej.2006.9.99. [DOI] [PubMed] [Google Scholar]
- 37.Jones L., Harold D., Williams J. Genetic evidence for the involvement of lipid metabolism in Alzheimer’s disease. Biochim. Biophys. Acta. 2010;1801:754–761. doi: 10.1016/j.bbalip.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Yun C.-H., Lee H.-Y., Lee S.K., Kim H., Seo H.S., Bang S., Kim S.E., Greve D.N., Au R., Shin C. Amyloid burden in obstructive sleep apnea. J. Alzheimer’s Dis. 2017;59:21–29. doi: 10.3233/JAD-161047. [DOI] [PubMed] [Google Scholar]
- 39.Ju Y.E.S., Finn M.B., Sutphen C.L., Herries E.M., Jerome G.M., Ladenson J.H., Crimmins D.L., Fagan A.M., Holtzman D.M. Obstructive sleep apnea decreases central nervous system-derived proteins in the cerebrospinal fluid. Ann. Neurol. 2016;80:154–159. doi: 10.1002/ana.24672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osorio R.S., Ayappa I., Mantua J., Gumb T., Varga A., Mooney A.M., Burschtin O.E., Taxin Z., During E., Spector N. The interaction between sleep-disordered breathing and apolipoprotein E genotype on cerebrospinal fluid biomarkers for Alzheimer’s disease in cognitively normal elderly individuals. Neurobiol. Aging. 2014;35:1318–1324. doi: 10.1016/j.neurobiolaging.2013.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bubu O.M., Pirraglia E., Andrade A.G., Sharma R.A., Gimenez-Badia S., Umasabor-Bubu O.Q., Hogan M.M., Shim A.M., Mukhtar F., Sharma N. Obstructive sleep apnea and longitudinal Alzheimer’s disease biomarker changes. Sleep. 2019;42:zsz048. doi: 10.1093/sleep/zsz048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liguori C., Mercuri N.B., Izzi F., Romigi A., Cordella A., Sancesario G., Placidi F. Obstructive sleep apnea is associated with early but possibly modifiable Alzheimer’s disease biomarkers changes. Sleep. 2017;40:zsx011. doi: 10.1093/sleep/zsx011. [DOI] [PubMed] [Google Scholar]
- 43.Maimon N., Hanly P.J. Does snoring intensity correlate with the severity of obstructive sleep apnea? J. Clin. Sleep Med. 2010;6:475–478. [PMC free article] [PubMed] [Google Scholar]
- 44.Folstein M.F., Folstein S.E., McHugh P.R. “Mini-mental state”. J. Psychiatr. Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z., Gerstein M., Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Audic S., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 48.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
- 49.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong H.K., Gim J.A., Yeo S.H., Kim H.S. Integrated late onset Alzheimer’s disease (LOAD) susceptibility genes: Cholesterol metabolism and trafficking perspectives. Gene. 2017;597:10–16. doi: 10.1016/j.gene.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Payao S.L., Goncalves G.M., de Labio R.W., Horiguchi L., Mizumoto I., Rasmussen L.T., de Souza Pinhel M.A., Silva Souza D.R., Bechara M.D., Chen E., et al. Association of interleukin 1beta polymorphisms and haplotypes with Alzheimer’s disease. J. Neuroimmunol. 2012;247:59–62. doi: 10.1016/j.jneuroim.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 53.Zhong A., Xiong X., Shi M., Xu H. Roles of interleukin (IL)-6 gene polymorphisms, serum IL-6 levels, and treatment in obstructive sleep apnea: A meta-analysis. Sleep Breath. 2016;20:719–731. doi: 10.1007/s11325-015-1288-6. [DOI] [PubMed] [Google Scholar]
- 54.Chuang L.P., Chen N.H., Lin Y., Ko W.S., Pang J.H. Increased MCP-1 gene expression in monocytes of severe OSA patients and under intermittent hypoxia. Sleep Breath. 2016;20:425–433. doi: 10.1007/s11325-015-1252-5. [DOI] [PubMed] [Google Scholar]
- 55.Honma T., Hatta K., Hitomi Y., Kambayashi Y., Hibino Y., Konoshita T., Nakamura H. Increased systemic inflammatory interleukin-1ss and interleukin-6 during agitation as predictors of Alzheimer’s disease. Int. J. Geriatr. Psychiatry. 2013;28:233–241. doi: 10.1002/gps.3816. [DOI] [PubMed] [Google Scholar]
- 56.Galimberti D., Fenoglio C., Lovati C., Venturelli E., Guidi I., Corra B., Scalabrini D., Clerici F., Mariani C., Bresolin N., et al. Serum MCP-1 levels are increased in mild cognitive impairment and mild Alzheimer’s disease. Neurobiol. Aging. 2006;27:1763–1768. doi: 10.1016/j.neurobiolaging.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 57.Tan Z.S., Beiser A.S., Vasan R.S., Roubenoff R., Dinarello C.A., Harris T.B., Benjamin E.J., Au R., Kiel D.P., Wolf P.A., et al. Inflammatory markers and the risk of Alzheimer disease: The Framingham Study. Neurology. 2007;68:1902–1908. doi: 10.1212/01.wnl.0000263217.36439.da. [DOI] [PubMed] [Google Scholar]
- 58.Sattlecker M., Khondoker M., Proitsi P., Williams S., Soininen H., Kloszewska I., Mecocci P., Tsolaki M., Vellas B., Lovestone S., et al. Longitudinal Protein Changes in Blood Plasma Associated with the Rate of Cognitive Decline in Alzheimer’s Disease. J. Alzheimer’s Dis. 2016;49:1105–1114. doi: 10.3233/JAD-140669. [DOI] [PubMed] [Google Scholar]
- 59.Pennisi M., Crupi R., Di Paola R., Ontario M.L., Bella R., Calabrese E.J., Crea R., Cuzzocrea S., Calabrese V. Inflammasomes, hormesis, and antioxidants in neuroinflammation: Role of NRLP3 in Alzheimer disease. J. Neurosci. Res. 2017;95:1360–1372. doi: 10.1002/jnr.23986. [DOI] [PubMed] [Google Scholar]
- 60.Emamian F., Khazaie H., Tahmasian M., Leschziner G.D., Morrell M.J., Hsiung G.Y., Rosenzweig I., Sepehry A.A. The Association Between Obstructive Sleep Apnea and Alzheimer’s Disease: A Meta-Analysis Perspective. Front. Aging Neurosci. 2016;8:78. doi: 10.3389/fnagi.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ancoli-Israel S., Klauber M.R., Butters N., Parker L., Kripke D.F. Dementia in Institutionalized Elderly: Relation to Sleep Apnea. J. Am. Geriatr. Soc. 1991;39:258–263. doi: 10.1111/j.1532-5415.1991.tb01647.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.