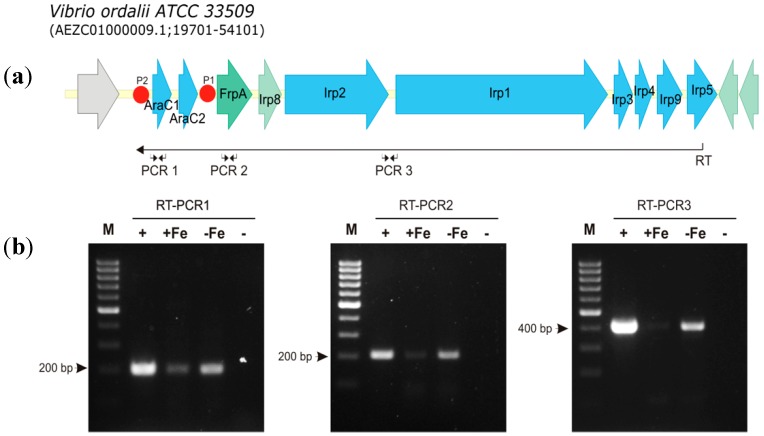
Figure 3.
Transcriptional organization of the gene cluster putatively encoding biosynthesis and transport of siderophore piscibactin in V. ordalii. (a) The predicted gene functions are: biosynthesis, genes irp1, irp2, irp3, irp4, irp5 and irp9; outer membrane receptor, frpA; transcriptional regulators, araC1 and araC2; and inner membrane exporter of putative siderophore, irp8. Predicted promoters P1 and P2 containing Fur boxes are indicated by red dots. RT denotes the location of primer used in retrotranscriptase reaction while PCR 1, PCR 2 and PCR 3 indicate location of primers for detection of cDNA from piscibactin gene cluster. (b) results of three RT-PCR reactions designed to analyze the transcription of the irp gene cluster. Primer marked as RT, targeted to the 3′-end of irp5 gene, was used to obtain a cDNA that spanned from irp5 to araC1. This cDNA was then used as template for three PCR reactions targeted within araC1 (RT-PCR1), frpA (RT-PCR2) and between irp2 3’-end and irp1 5’-end (RT-PCR3). M, size marker from 100 to 1000 bp. Negative controls (-) are RT-PCR reactions lacking reverse transcriptase. Positive controls (+) are PCR reactions using chromosomal DNA as template, +Fe: RT-PCR performed with cells grown under iron excess (TSB-1 + FeCl3 20 µM); -Fe: RT-PCR performed with cells grown under iron limitation (TSB-1 + 2,2′-dipyridyl 60 µM).