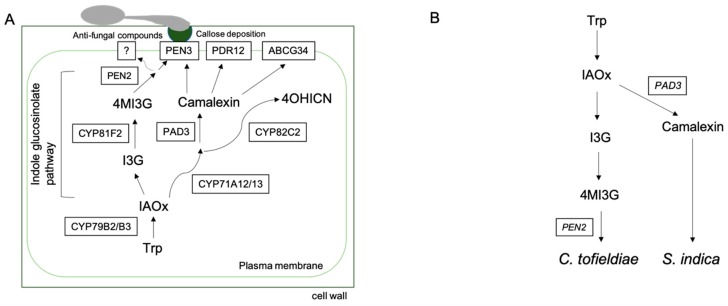
Figure 1.
Overview of the tryptophan-derived metabolite pathway required for plant–microbe interactions. (A) Tryptophan (Trp) is converted to indole-3-acet aldoxime (IAOx) by the cytochrome P450 members CYP79B2 and CYP79B3. The indole glucosinolate pathway, the camalexin pathway, and the 4-hydroxyindole-3-carbonitrile (4OHICN) pathway are reportedly required for defense against pathogens. In the indole glucosinolate pathway, indol-3- ylmethyl glucosinolate (I3G) is converted to 4-methoxyindol-3-ylmethylglucosinolate (4MI3G) by the cytochrome P450 CYP81F2. 4MI3G is then hydrolyzed by the atypical myrosinase PEN2, and the resulting compounds are transported by the ABC transporter PEN3. In contrast, the weaker phenotype of pen3 compared to pen2 plants against non-adapted Colletotrichum fungi suggests that additional ABC transporters may participate in the export of metabolites generated by PEN2 [19]. In the camalexin pathway, both cytochrome P450 CYP71A12/CYP71A13 and PAD3 (CYP71B15) generate the phytoalexin camalexin. Camalexin is reportedly transported by PEN3, PDR12, and PDR6/ABCG34 [25,35]. CYP71A12/CYP71A13 and cytochrome P450 CYP82C2 are required for the generation of 4OHICN, which is required for defense against both fungal and bacterial pathogens. (B) The indole glucosinolate pathway is required for beneficial interactions with the root endophyte Colletotrichum tofieldiae (Ascomycete). The camalexin pathway is required for beneficial interactions with the root endophyte Serendipita indica (Basidiomycete).