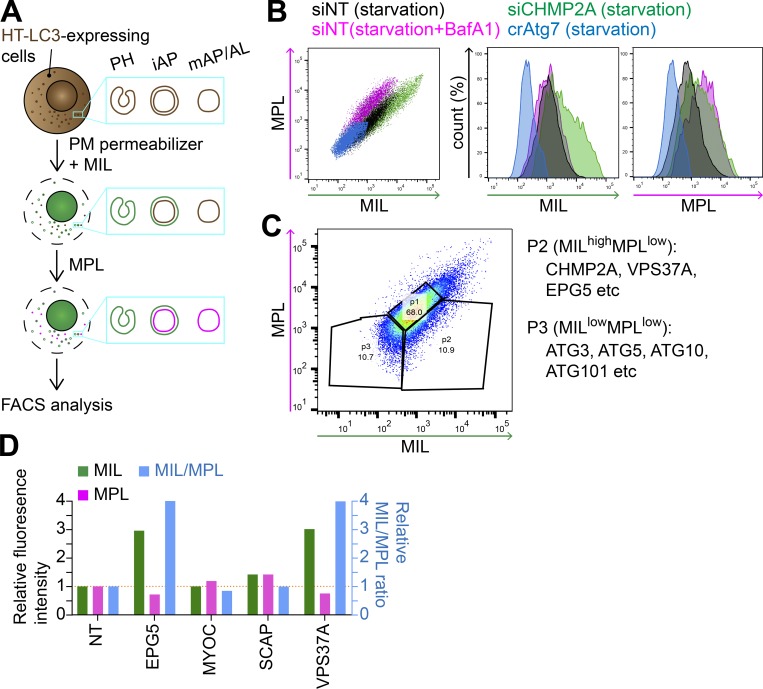
Figure 1.
An HT-LC3 assay–based CRISPR screen identifies VPS37A and EPG5 as potential regulators of autophagosome completion. (A) Schematic strategy of the flow cytometry–based HT-LC3 autophagosome completion assay. HT-LC3–expressing cells are permeabilized and sequentially incubated with AF488-conjugated MILs followed by TMR-conjugated MPLs to label autophagosomal membrane-unsequestered and sequestered HT-LC3-II, respectively. The fluorescence intensities of AF488 and TMR are then analyzed by FACS. iAP, immature autophagosome; mAP/AL, mature autophagosome/autolysosome; PH, phagophore; PM, plasma membrane. (B) HT-LC3–expressing U-2 OS cells transiently transfected with nontargeting siRNA (siNT) or CHMP2A siRNA (siCHMP2A) for 48 h, and HT-LC3–expressing crAtg7 cells were starved in the presence or absence of 100 nM BafA1 for 3 h and subjected to the HT-LC3 assay followed by FACS analysis. (C) The human Brunello lentiviral CRISPR pooled library was used to transduce HT-LC3–expressing U-2 OS cells. 7 d after the transduction, the resultant cells were starved for 3 h and subjected to the HT-LC3 assay followed by FACS analysis. The MILhighMPLlow and MILlowMPLlow populations indicated by the ROIs were sorted and analyzed by next-generation sequencing. Data shown are representative of four independent screens. (D) The selected hits from P2 of the primary screen in C were subjected to secondary screening. Three lentiviruses encoding different gRNAs against a gene were pooled and transduced into the HT-LC3–expressing U-2 OS cell for 2 d. Cells were then selected with 1 µg/ml puromycin for 3 d and then cultured at least another 3 d in complete medium. The resultant stable transfectants were starved in the presence of 100 nM BafA1 for 3 h and subjected to the HT-LC3 assay followed by FACS analysis. The AF488 and TMR intensities relative to those of control crNT cells are shown (n = 1).