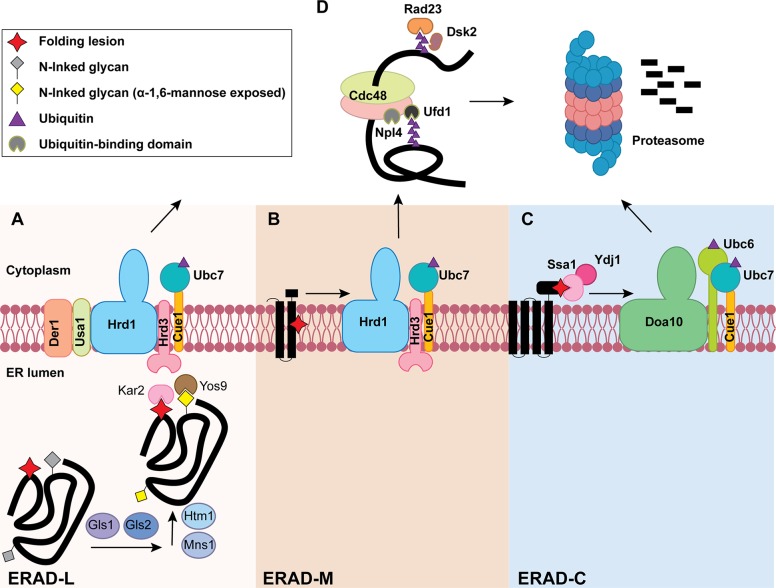
Figure 1.
The three branches of the ERAD pathway in yeast. Lower panels in A, B, and C define the different steps during ERAD-L, ERAD-M, and ERAD-C, respectively. (A) ERAD-L substrates containing lumenal folding lesions (red star) and N-linked glycans (gray diamond) are recognized and processed by an enzyme cascade to generate an ERAD-targeting glycan (yellow diamond). Chaperones (e.g., Kar2) and lectins (e.g., Yos9) capture the substrate for transfer to the Hrd1 complex for retrotranslocation-coupled ubiquitination (purple triangle). (B) ERAD-M substrates containing a membrane-embedded folding lesion (red star) are recognized and ubiquitinated by the Hrd1 complex. (C) ERAD-C substrates containing cytosolic folding lesions are instead recognized by cytosolic chaperones (e.g., Ydj1/Hsp40 and Ssa1/Hsp70), which bridge the Doa10 ubiquitin ligase to an ERAD substrate. The three ERAD branches converge at a Cdc48-complex–dependent retrotranslocation step (D, top). The Cdc48 complex also contains Ufd1/Npl4, which interacts with ubiquitin (purple triangle). Following retrotranslocation, substrates are escorted to the 26S proteasome for degradation with the help of the Ras23 and Dsk2 shuttling factors.