Abstract
Motivation
Searching for precise terms and terminological definitions in the biomedical data space is problematic, as researchers find overlapping, closely related and even equivalent concepts in a single or multiple ontologies. Search engines that retrieve ontological resources often suggest an extensive list of search results for a given input term, which leads to the tedious task of selecting the best-fit ontological resource (class or property) for the input term and reduces user confidence in the retrieval engines. A systematic evaluation of these search engines is necessary to understand their strengths and weaknesses in different search requirements.
Result
We have implemented seven comparable Information Retrieval ranking algorithms to search through ontologies and compared them against four search engines for ontologies. Free-text queries have been performed, the outcomes have been judged by experts and the ranking algorithms and search engines have been evaluated against the expert-based ground truth (GT). In addition, we propose a probabilistic GT that is developed automatically to provide deeper insights and confidence to the expert-based GT as well as evaluating a broader range of search queries.
Conclusion
The main outcome of this work is the identification of key search factors for biomedical ontologies together with search requirements and a set of recommendations that will help biomedical experts and ontology engineers to select the best-suited retrieval mechanism in their search scenarios. We expect that this evaluation will allow researchers and practitioners to apply the current search techniques more reliably and that it will help them to select the right solution for their daily work.
Availability
The source code (of seven ranking algorithms), ground truths and experimental results are available at https://github.com/danielapoliveira/bioont-search-benchmark
Keywords: information retrieval, ranking algorithms, ontology, healthcare and life sciences, semantic Web, linked data
Introduction
The biomedical domain has a long history of using formal vocabularies, terminologies, codes and ontologies to describe data sets ranging from genetics, molecular or chemical domains to patients receiving medical care [1, 2]. This domain has been one of the early adopters of Artificial Intelligence (AI), Semantic Web and recently introduced linked and big data initiatives, resulting in the development of several biomedical repositories and ontologies.
Biomedical ontologies are typically large [3], covering thousands of concepts represented by classes (e.g. the Gene Ontology [4] has almost 50 K classes). Most ontologies in this domain use a rich vocabulary in labels, synonyms and textual definitions associated with classes and properties [5]. For example, the class http://purl.obolibrary.org/obo/GO_1905294 has (1) a preferred label: positive regulation of neural crest cell differentiation; (2) a textual definition: any process that activates or increases the frequency, rate or extent of neural crest cell differentiation; and (3) synonyms: upregulation of neural crest cell differentiation; upregulation of neural crest cell differentiation; upregulation of neural crest cell differentiation; and activation of neural crest cell differentiation.
A key barrier for a data publisher, however, is to find the right set of ontologies, terminologies or vocabularies to annotate entities in data sets. Often the search results over the ontology repositories are overwhelming, with dozens of synonyms matching in different ontologies, as well as a common disagreement between search engines in the ranking of ontological resources in their search results. However, various data publishing platforms (in the biomedical context) and infrastructures advocate that if a database entity is described using a precise ontological resource, it eventually leads to efficient linking and querying over published data sets [6, 7]. Owing to different naming conventions, textual descriptions, synonyms and granularity of the biomedical entities, it is an open research problem to precisely identify an ontological resource, which best describes a given concept.
The difficulty of finding ontological resources (i.e. classes and properties) for a given set of words has direct and indirect consequences (i) newly introduced repositories often develop proprietary schemas (including terminologies and codes) that fit well for a particular use case, developing an ontology from scratch and hampering the reuse of well-established ontologies; (ii) a recent study about Linked Open Data [LOD (http://lod-cloud.net/)] suggests that data sets based on closed and/or proprietary schemas end up being isolated with fewer incoming links, thus, impeding the main purpose of using linking data across several co-related facilities [7]; and finally, (iii) loosely annotated data sets—especially in the biomedical domain—severely affect the horizontal layers (data curation, alignment, querying, entity disambiguation, etc.) of a data integration solution, resulting in weak data interoperability.
This research is motivated by the needs of the BIOOPENER project (http://bioopenerproject.insight-centre.org), which aims to link cancer and biomedical data repositories by providing interlinking and querying mechanisms to understand cancer progression. The BIOOPENER project needs to find the most appropriate ontological classes and properties to describe thousands of data entities independently created in cancer-related repositories. These repositories are not linked to each other and after converting them to RDF format, the free-text in the database needs to be annotated with ontological resources. However, to the best of our knowledge, no systematic study has been conducted to compare state-of-the-art algorithms and search engines in the domain of biomedical ontologies to conclude their reliability and to understand what search scenarios motivate the use of each algorithm or service to choose the adequate one for the project’s needs.
In this article, we extend our initial work [8] by testing state-of-the-art Information Retrieval (IR) algorithms, ontology ranking approaches and established search engines for searching and ranking biomedical ontologies. The algorithms and search applications/engines are tested by searching a defined set of queries obtained from a cancer genomics scenario. Using these queries, we established a ground truth (GT) by asking 10 biomedical and ontology engineering experts to manually rank the search results for each query. As building a manual GT is an expensive process, we developed an automated probabilistic ground truth (PGT). The PGT led to a better understanding of the GT and allowed the expansion of the ontology collection and the number of queries tested. We then compared both ground truths (GT and PGT) with the results of the algorithms and applications obtained by using Precision@k, Average Precision@k, Mean Average Precision (MAP) and Normalized Discounted Cumulative Gain (NDCG). This evaluation provided the necessary knowledge to explore the advantages and disadvantages of using each search engine and ranking algorithm.
We start the article by discussing relevant related work, followed by the description of two biomedical search services [BioPortal and Ontology Lookup Service (OLS)], a search library (Solr) and an annotation tool (Zooma) and then seven ranking algorithms. We present the experimental set-up by describing the ontology and queries used and by explaining the construction of the GT and PGT. We provide an analysis and discussion of our approach and results. Finally, we present our conclusions in regards to the algorithms and applications tested and give an overall view of the state of the art of searching biomedical ontologies with query words.
Related work
IR approaches have been successful in finding and ranking relevant documents. In the Web environment, IR search engines are primarily keyword-based and analyse the relevance of a document using content-based or graph-based methods. Early approaches to query search and ranking focused on entities of different types, which are present in Wikipedia [9]. Similarly, classic named-entity recognition (NER) approaches aim to find information about a given set of entities in a text. Often, NER-based techniques find results for generic and limited sets of entity types (e.g. Person, Organization, Address). Semantic Search engines have benefited from well-established IR methods. For instance, Swoogle (which was initially developed to rank ontologies only) [10], Sindice.com [11], Watson [12] or Yars2 [13] allow searching of ontology resources through user queries. The ranking in these search engines follows traditional link-based ranking methods, in particular, adapted versions of the PageRank algorithm [14], where links from one source of information to another are regarded as a ‘positive vote’ from the former to the latter. Falcons [15] is a popularity-based scheme to rank concepts and ontologies. However, most of these strategies focus on ranking instances but are not as effective when ranking classes or properties in ontologies.
Ranking ontological resources can be based on different criteria [16], for example, how well an ontology meets the requirements of certain evaluation tests [17] or on methods to evaluate general properties of an ontology based on some requirement [18]. However, only limited work has been proposed to rank resources based on user posed queries. AKTiveRank [19] is a system that uses structural metrics [i.e. Semantic Similarity, Betweenness, Density and Class Match Measure (CMM)] to evaluate different representational aspects of an ontology and calculates its ranking in relation to a set of search queries specified by a user. BioPortal also developed a tool [Ontology Recommender (https://bioportal.bioontology.org/recommender)] that, from a set of keywords or a text, tries to return the lowest number of ontologies the covers the input.
IR algorithms have been successfully applied in a few open-source indexing and search engines, such as Lucene (http://lucene.apache.org/), Solr (http://lucene.apache.org/solr) and ElasticSearch (https://www.elastic.co/). These applications include Application programming interfaces (API) to provide an easy implementation and fast search. The user has control over most aspects of the inner-workings of these applications and can adapt them to serve specific needs, e.g. ontology search.
General search services and algorithms have been developed for linked data applications, for instance, the Linked Open Vocabularies (LOV) (http://lov.okfn.org/dataset/lov/), YAGO [20], OntoKhoj [21], OntoSearch [22] or OntoSelect [23]. However, none of them is designed specifically for the biomedical domain; therefore, some of them (e.g. LOV and YAGO) index none of the available biomedical ontologies (LOV and YAGO), and others are not available any more (OntoSelect, OntoSearch and OntoKhoj). OntoSelect provided an evaluation methodology [24] by creating a benchmark that associated topics from Wikipedia pages with ontologies and then compared the retrieval results of OntoSelect with Swoogle. However, the authors concluded that, on average, OntoSelect did not perform better than Swoogle.
The biomedical community has made a significant effort to develop services such as BioPortal [25] and the OLS [26] for searching and applying ontological resources. However, they often suggest large, vague or loose search results for a given query. Searching for the right concept in the most appropriate ontology is, therefore, a strenuous task, as a significant number of available ontologies exist, in the same or in closely related domains, that describe overlapping, closely related or the exact same concept.
OntoCAT [27] provides uniform access for search across different public online repositories (BioPortal and OLS) but also allows the inclusion of local ontology files in standard OWL or OBO formats. This software is available as an R package [28] and is an easy method to programmatically search and integrate ontologies from different origins in the R environment.
Ontology search: applications and algorithms
Searching applications
We analysed and compared two biomedical repositories with integrated online search services, a local stand-alone search engine and an annotation tool. The online applications have APIs publicly available that were searched in the version available on the 19 December 2017. The following sections describe the ontology search applications:
BioPortal
BioPortal is a repository containing both open-access and licenced biomedical ontologies and terminologies. Since its inception, the BioPortal library has grown substantially, from 72 ontologies in 2008 to over 200 in 2011 and, in 2017, already indexes >500 ontologies. Besides being a repository for biomedical ontologies, BioPortal includes other resources and services. One of them is providing a search mechanism to find ontologies or ontology resources through keyword search. This search usually returns several matches, and the results are ranked by the popularity (i.e. number of visits), in BioPortal, of their source ontology.
Solr
Solr is a platform that extends the Apache Lucene search library for full-text indexing and search. One of Solr’s major features is a REST-like API for easy integration with any programming language. The Lucene engine used by Solr scores documents using a combination of the Boolean model and a Vector Space Model (VSM) algorithm. First, it uses the Boolean model to narrow down the number of documents it needs to score and then uses the VSM to attribute a final score to a document in relation to a user’s query.
Ontology Lookup Service
The OLS is a repository for biomedical ontologies. As of January 2018, OLS has 206 ontologies and provides a search mechanism to match query words with ontological concepts. This search uses Apache Solr to index ontologies, but it applies specific boosts to some of the results, such as label or ID exact matches.
Zooma
Zooma (http://www.ebi.ac.uk/spot/zooma/) provides mappings between a free-text input and a curated repository of annotation knowledge. This repository contains the annotations that were manually associated with data from sources such as the Expression Atlas [29] and the Genome-Wide Association Studies (GWASs) catalogue [30]. When no mappings are found in the curated data repository, OLS search is used instead to increase coverage.
Ranking algorithms
We implemented seven commonly used ranking algorithms for documents and adapted them to, given a free-text query, rank resources in a collection of ontologies. For content-based algorithms [i.e. term frequency–inverse term frequency (tf-idf), BM25, VSM and CMM), instead of using words as the base unit, we considered a resource r (class or property) in the ontology as the measuring unit. A resource is matched to a query if any of the query words exist in the values for the label, synonyms or description. When we wish to retrieve only exact matches, the query words have to be strictly the same as the value matched from the label, synonyms or description of a resource. The graph-based models (PageRank and Semantic Similarity) do not consider properties, only classes. However, <1% of all resources in the collection are properties.
Table 1 lists the formal notations applied in the description of the algorithms. The following sections describe the algorithms with their adaptation for ranking ontologies.
Table 1.
Notation used
| Variable | Description |
|---|---|
| Ontology collection | |
| N | Number of ontologies in |
| O | An ontology: |
| R | Collection of all resources (i.e. classes and properties) with R ∈ O |
| r | A resource: r ∈ O and r ∈ R |
| Q | Query string |
| qi | Query word i of Q |
| σO | Set of matched resources r for Q in O |
| σO(qi) | Set of matched resources r for qi in O: ∀ri ∈ σO, ri ∈ O and ri matches qi |
Boolean model
The standard Boolean model is based on Boolean algebra, where a query is viewed as a Boolean expression. Therefore, for a set of ontologies and queries, the retrieval is binary and based on whether the retrieved results contain the query words.
Term frequency–inverse term frequency
tf-idf [31] quantifies how important a term is in an ontology by analysing the frequency of the term in the resources of that ontology and in the overall collection of ontologies.
| (1) |
Here tf(r, O) is the term frequency of r in O obtained by dividing the frequency of r by the maximum frequency of any resource rj ∈ O. The inverse document frequency is a measure of commonality of a resource across the collection. It is obtained by dividing the total number of ontologies in the collection, N, by the number of ontologies containing the resource r and then computing the logarithm of that quotient. The final tf-idf of r is the product of the tf and the idf.
BM25
BM25 [32] is a weighting scheme that takes into account, not only term frequency but also ontology size without introducing too many additional parameters in relation to tf-idf. Usually, the BM25 score is computed for ∀qi ∈ Q, but, to tailor this statistic for ontology ranking, we compute the sum of the score of each rj ∈ σO(qi) for each query term qi. Therefore, given a resource r ∈ σO(qi), with a value (e.g. label) containing the words r1,…, rn, the BM25 score of the ontology O is computed by:
| (2) |
where tf(rj, O) is term frequency for the matched resource rj in the ontology O, and is the inverse document frequency of the resource rj ∈ σO(qi). |O| is the total number of resources (i.e. 3 ×|axioms|) in the ontology, and avgol is the average ontology size in the ontology collection. k and b are free parameters, usually chosen, in absence of an advanced optimization, as k ∈ [1.2, 2.0] and b = 0.75. For the current implementation, we used k = 2.0, b = 0.75.
Vector Space Model
A VSM [33] assumes that ontology resources and queries can be represented by the same type of vector. Non-binary weights are assigned to indexed terms, usually using weighting schemes such as tf-idf. The degree of similarity between query and ontology resources is calculated by comparing the vectors that represent the query and each ontology resource. The VSM score was calculated as follows:
| (3) |
Here, w(qi, O) and w(qi, Q) are the weights of qi in the ontology O and query Q, respectively. |O| is the ontology vector norm, and |Q| is the query vector norm. For this implementation, we consider tf-idf as the vector weight. Therefore, the similarity of an ontology to query Q is computed as:
| (4) |
PageRank
PageRank [34] is an iterative method to analyse links, and it was adapted to assign a numerical score to each ontology in a set of ontologies. This implementation considers ontologies as nodes and owl: imports (imports of other ontologies into the current ontology, i.e. outlinks) as edges. In each successful iteration, the score of the ontology O is determined as the sum of the PageRank score of the previous iterations of all the ontologies that import ontology O divided by their number of outlinks. For the kth iteration, the PageRank score of ontology O is given by:
| (5) |
Here, are ontologies in the collection that have no outlinks. All nodes are initialized with an equal score (i.e. , where N is the total number of ontologies in before the first iteration). In the experimental evaluation, we set the damping factor d equal to 0.85 (common practice).
Ontologies with no owl: imports statement can still reuse classes from other ontologies following the MIREOT [35] guidelines for referring external terms from a target ontology. In our experiment, whenever these references existed within the ontology classes, an owl:imports statement was introduced to identify the link between the two ontologies.
Class Match Measure
CMM [19] calculates the coverage score of an ontology in relation to a set of given queries. Despite not ranking each query individually, this algorithm represents the type of search one could expect from a user that requires the lowest number of ontologies to cover all the queries in their search.
The CMM algorithm looks for exact and partial matches and scores an ontology depending on the number of matches. A higher number of matches mean a higher CMM score. The score for an ontology is computed as:
| (6) |
where is the final score for CMM, is the exact match measure and is the partial match measure for the ontology O with respect to the set of queries . α and β are the exact matching and partial matching weight factors, respectively. Exact matching is favoured over partial matching, and therefore, α > β. Here, α = 0.6 and β = 0.4.
| (7) |
φ(r, Q) counts the number of exacts matches, and ψ(r.Q) counts the number of partial matches. and sum the number of matches (exact and partial, respectively) that exist in every ontology for a set of queries.
Semantic Similarity Measure
The Semantic Similarity Measure (SSM) [19] applied here takes advantage of the ontological graph structure to calculate how close resources are in the ontology structure.
scoreSSM(O, Q) is the SSM score of ontology O for a given query Q. It is a collective measure of the shortest path lengths for all classes that match the query string.
| (8) |
Summary
Table 2 presents a summary of the characteristics of the algorithms. The table shows: (1) the main scoring mechanism of each algorithm, (2) if the algorithm attributes a global score to the ontology or scores each resource in the ontology individually, (3) if there is any distinction between partial matches and exact matches (yes) or if they are treated equally (no) and, finally, (4) a summary of the conclusions presented in [8].
Table 2.
Summary of IR algorithms
| Algorithm | Scoring | Global | WPM | Remarks |
|---|---|---|---|---|
| tf-idf | Term frequency | No | No | Frequent resources in the collection have a low score. In ontologies, a common term does not necessarily mean less relevant. Frequent terms can be a product of reuse by other ontologies |
| BM25 | Term frequency | Yes | No | Suffers from the same issue has tf-idf, but the cumulative score ranks domain ontologies higher |
| VSM | Vector similarity | No | No | Uses tf-idf to weight vectors and also considers the tf-idf of the query, aggravating the tf-idf drawback |
| PageRank | Links between ontologies | Yes | No | Ranks based on popularity, which may lead to popular but less relevant resources, being ranked higher |
| CMM | Coverage of the set of queries | Yes | Yes | Ontologies with a large number of partial matches will be scored higher than ontologies with few exact matches |
| SMM | Closeness between ontological resources | Yes | No | Although this algorithm can be useful when considering similarity among the matched resources of two or more query terms of a multi-keyword query, it performs poorly on single-word queries |
Note: Scoring summarizes the main scoring method of the algorithm. Global indicates if the score attributed by the algorithm is per resource or per ontology. WPM (weights partial matches) shows if the ontology distinguishes between partial and exact matches.
Evaluation: ontology search applications and algorithms
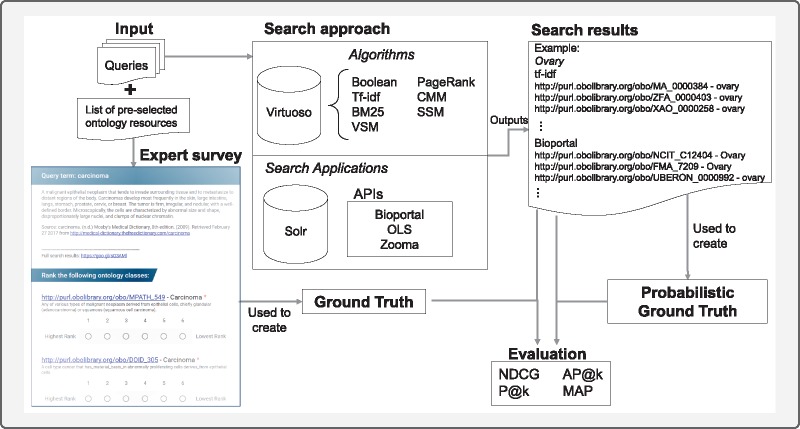
This work was divided into two separate but comparable analyses that give a complete overview of the performance of the chosen applications and algorithms. The first approach evaluated the results against a GT obtained from a questionnaire answered by experts. The second analysis was based on an automated PGT obtained from the consensus between the algorithms and four search applications. Figure 1 illustrates how the search process progresses from the queries to the different algorithms/applications and presents an example of the search results. The figure shows the evaluation process starting from the creation of the GT and the PGT and their comparison with the search results, and finally obtaining the evaluation results. The following subsections explain the processes common to both analysis and then detail how both ground truths were obtained and validated.
Figure 1.
Evaluation workflow: from input search queries to evaluation results.
Ontology loading
The ontology resources were stored in a Virtuoso database by treating the ontologies as sets of triples. For example, the following triple of the HP ontology was loaded into the virtuoso database: obo: HP_0006719 rdf: type owl: Class (Prefixes: obo: <http://purl.obolibrary.org/obo/>; rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>; owl: < http://www.w3.org/2002/07/owl#>). In total, the ontologies define around 20 M triples and 645 K distinct classes (subjects of rdf: type owl: Class). In Solr, ontologies were loaded using the method provided by the OLS development team (https://github.com/EBISPOT/OLS/tree/master/ols-apps/ols-solr-app), which uses the owlapi Java API [36] to manipulate the provided ontologies formatted with the Web Ontology Language (OWL) (https://www.w3.org/OWL).
Performance metrics
The evaluation of the algorithms and search applications was based on four metrics: Precision at k, Average Precision at k, MAP and Normalised NDCG. These metrics evaluate the results against the GT and the PGT. Both ground truths have a number of defined relevant search results that vary for each query. For example, in the GT, the query ‘MYH7’ only had one relevant result while the query ‘Ovary’ had five. In terms of metrics, this difference means that, with a fixed k = 3, if a search of the query ‘MYH7’ returned more than one result, the precision would be lower than expected, even if the first result was the correct one. Therefore, instead of choosing a fixed cut-off, the k is chosen independently for each query and each GT depending on the number of search results present in the respective GT. Therefore, the query ‘MYH7’ was evaluated with a k = 1, while the query ‘Ovary’ had a k = 5. This adaptation evaluates if the algorithms and search application return all the relevant results in the first k positions.
The metrics were calculated as follows:
Precision@k (P@k)
| (9) |
Average Precision (AP@k) for a query Q is defined as:
| (10) |
where rel(ri) is 1 if ri is a relevant resource for the query Q and 0 otherwise, P@i is the precision at i and k is the cut-off value.
MAP is the mean of AP(Q) over all queries and is calculated as:
| (11) |
NDCG is a standard evaluation measure of ranking quality that allows graded relevance instead of the traditional binary relevance. NDCG involves a discount function to weight the rank for penalizing relevant resources that appear in a low position in the search result. The Discounted Cumulative Gain (DCG) is calculated with:
| (12) |
The NDCG is the quotient between the obtained DCG value and the ideal DCG value (iDCG). The iDCG is calculated by sorting the results from most to least relevant. In the context of this work, the iDCG is the DCG of the GT ordering.
Building expert-based GT
The GT was established with a study (The questionnaire is available at https://goo.gl/pQUvte) involving 10 experts that were asked to rank the ontology resources matched with the 10 query terms.
Ontology collection and search queries
A collection of 23 ontologies (Table 3) representative of different domains in the biomedical field was used. The domains chosen range from chemical compounds to diseases or phenotypes, among others. The collection also includes different species such as mouse and zebrafish. The set of ontologies has some of the most popular and freely accessible biomedical ontologies, with more than half of them included in the top 50 of BioPortal’s most visited ontologies (as of December 2017).
Table 3.
Ontologies used in this benchmark with name, acronym, number of triples and reference
| Name | Acronym | # Triples |
|---|---|---|
| Chemical Entities of Biological Interest Ontology [37] | ChEBI | 8187078 |
| Cell Ontology [38] | CL | 69796 |
| Human Disease Ontology [39] | DOID | 203125 |
| The Drug Ontology [40] | DRON | 138898 |
| EMBRACE Data And Methods [41] | EDAM | 33300 |
| Experimental Factor Ontology [42] | EFO | 469954 |
| Foundational Model of Anatomy [43] | FMA | 612982 |
| Gene Ontology [4] | GO | 1575776 |
| Human Phenotype Ontology [44] | HP | 350017 |
| Mouse Adult Gross Anatomy Ontology [45] | MA | 25523 |
| Mammalian Phenotype Ontology [46] | MP | 335821 |
| Mouse Pathology Ontology [47] | MPATH | 11992 |
| Neuro Behavior Ontology [48] | NBO | 10376 |
| National Cancer Institute Thesaurus [49] | NCIT | 5784846 |
| Ontology of Adverse Events [50] | OAE | 54334 |
| Ontology of Genes and Genomes [51] | OGG | 1211539 |
| Phenotypic Quality Ontology [52] | PATO | 31644 |
| Plant Ontology [53] | PO | 59932 |
| Uber Anatomy Ontology [54] | UBERON | 690529 |
| Vertebrate Trait Ontology [55] | VT | 44183 |
| Caenorhabditis elegans Phenotype Vocabulary [56] | WPhenotype | 31991 |
| Xenopus Anatomy and Development Ontology [57] | XAO | 40611 |
| Zebrafish Anatomy and Development Ontology [58] | ZFA | 82964 |
The searches tried to match each query with all ontological resources available in each platform, i.e. online applications used their services and local tests used the Virtuoso database or local Solr server. When using the search applications, results that included ontologies outside this list were excluded. The search applications and the ranking algorithms were tested using a set of queries in the domain of ovarian cancer. Table 4 presents the 10 search terms chosen from the BIOOPENER project repositories and the abbreviations used in this article. Despite belonging to the domain of ovarian cancer, the selected terms represent different branches of the biomedical domain, i.e, the terms represent diseases, drugs, tumours, organs and genes.
Table 4.
Cancer-related queries and their number of search results on Google, BioPortal and OLS, in April 2017
| Query terms | Abbreviation | Type | BioPortal | OLS | |
|---|---|---|---|---|---|
| Ovary | Ovary | Organ | 25.400.000 | 29 | 1054 |
| MYH7 | MYH7 | Gene | 86.500 | 8 | 22 |
| Paclitaxel | Paclitaxel | Drug | 4.640.000 | 18 | 149 |
| Carcinoma | Carcinoma | Disease | 32.800.000 | 25 | 4025 |
| Carboplatin | Carboplatin | Drug | 2.710.000 | 19 | 212 |
| Ovarian teratoma | OT | Tumour | 434.000 | 18 | 1164 |
| Ovarian cystadenoma | OCys | Tumour | 148.000 | 18 | 1100 |
| Ovarian choriocarcinoma | OChor | Tumour | 317.000 | 20 | 1129 |
| Ovarian embryonal carcinoma | OEC | Tumour | 164.000 | 19 | 5069 |
| Ovarian mucinous adenocarcinoma | OMA | Tumour | 117.000 | 15 | 2235 |
The general frequency of the terms was assessed with a Google search, which showed that the queries ‘Carcinoma’ and ‘Ovary’ are the most popular results and queries related to disease names were less common. The query 'MYH7' is related to a very specific gene name and, therefore, was the search with the least results. Finally, all the queries were used as input in BioPortal and OLS’ Web search, with the default parameters (search through all the ontologies and show exact and partial matches).
Experts
The experts were sourced from the IBM Research, USA; King Abdullah University of Science and Technology, Kingdom of Saudi Arabia; Maastricht University, The Netherlands; Medical University of Graz, Austria; Indian Institute of Technology (IIT) Bombay, India; Saarland University, Germany; Universite de Rennes 1, France; and the US National Library of Medicine, USA. The areas of expertise of these judges included knowledge engineering and the biomedical domain, with all of them having at least some experience in both domains.
To assess the level of experience in the biomedical and knowledge engineering fields, each expert was asked to rate his knowledge in a Likert scale of five levels, from ‘No Knowledge’ to ‘Expert Knowledge’. Table 5 shows that most experts have considered themselves to have a strong to medium knowledge in both domains apply or work with biomedical data. All of the judges have worked with ontologies, six worked specifically with biomedical ontologies and some have developed ontologies.
Table 5.
Level of self-accessed knowledge of the experts in the biomedical and knowledge engineering fields
| Expert | Biomedical | Works with BD | Produces BD | Applies BD | Knowledge engineering | Worked with Ont. | Developed a BmO |
|---|---|---|---|---|---|---|---|
| 1 | 5 | Yes | Yes | Yes | 2 | Yes | No |
| 2 | 3 | Yes | Yes | Yes | 5 | Yes | Yes |
| 3 | 5 | Yes | No | Yes | 5 | Yes | Yes |
| 4 | 4 | Yes | No | Yes | 5 | Yes | Yes |
| 5 | 5 | Yes | Yes | No | 4 | Yes | No |
| 6 | 4 | Yes | No | Yes | 5 | Yes | Yes |
| 7 | 5 | No | No | Yes | 3 | Yes | Yes |
| 8 | 4 | Yes | Yes | No | 3 | Yes | No |
| 9 | 5 | Yes | Yes | Yes | 5 | Yes | Yes |
| 10 | 5 | No | No | Yes | 5 | Yes | Yes |
| Average or ratio yes:no | 4.5 | 8:2 | 5:5 | 8:2 | 4.2 | 10:0 | 7:3 |
Note: BD = biomedical data; ont. = ontology; BmO is biomedical ontology. Bold numbers distinguish which values refer to the average.
Questionnaire
The experts were given a list of ontology classes to rank in relation to 10 queries (Table 4). These classes were obtained by searching BioPortal and OLS and also from the search results of the IR algorithms. All results were merged and taken out of order. The results presented to the judges were composed by the classes with labels that were an exact match of the query terms. However, the judges still had access to all the retrieved classes and could introduce classes they deemed relevant but were not shown. The questionnaire presented definitions from medical dictionaries to establish the search intention of each query. The definitions did not follow ontological definitions patterns. Their main goal was to lower the ambiguity of the queries and to provide some guidance to the judges. For each of the classes displayed in the questionnaire, we provided the class Uniform Resource Identifier (URI) preferred label and definition. The judge could rank the classes in a Likert-type scale with a number of options equal to the number of items to rank, with the first option corresponding to the best rank, plus the last rank reserved to mark the search term as ‘not-relevant’.
Validation
The questionnaire answers of each judge were evaluated with two different approaches:
Rank agreement considers the observed agreement between the ranks allocated to each search result.
Relevancy agreement analyses the results in terms of the observed agreement in a binary scale of relevant/not-relevant.
For each approach, the answers of each judge were compared in pairs, and the final result was obtained by averaging the pairwise agreement.
The answers were further analysed with a chi-square goodness-of-fit test [59], which is a non-parametric statistical test to determine if an observed value is significantly different than its theorized value. Therefore, this test was applied to assess the randomness of the expert’s answers. The null hypothesis considered was ‘each rank has an even number of answers’, which leads to the alternative hypothesis ‘the ranks are not equally chosen among experts’. This test was performed for each query with a significance level of 0.05.
Building PGT
To validate the GT as well as to include a more diverse set of queries and ontologies, we extended the approach used in [60] to build a PGT without involving human experts. In [60], the authors present an approach to calculate the probability that each document in a collection belongs to a possible GT by using the consensus from multiple systems as the reference. The main idea in [60] is that a probabilistic GT is equivalent to an unknown GT and contains the probabilities of each document (δi) that appears in the search results to belong to a real GT. These probabilities are calculated with the following:
| (13) |
where s is the set of systems being tested, and Sk(δi) represents the search result of document δi by system Sk. The authors of [60] consider the result to be binary, i.e. the document δi is either present or absent of the search results of system Sk. By using these probabilities, the authors then calculate a probabilistic precision and recall. For the purpose of our work, however, a comparison with the GT was necessary. Therefore, we developed an extended PGT that can be compared with the expert-based GT using the performance measures.
Extending and building the PGT
To extend the original approach to the ontology context, instead of using ontologies as documents, each ontology resource was considered the measuring unit. This adaptation allows the ranking of several resources matched in the same ontology.
Using the principle of the DCG, a ranking approach was added to the creation of the PGT to obtain a non-binary classification of each resource. The method used a penalty measure—in a logarithmic scale [61]—for search results that appear lower on the list. The penalty is proportional to their position p. Considering all search results relevant, the discount metric (DM) is obtained with:
| (14) |
The probability of each ontology resource belonging to a possible GT is:
| (15) |
Resources with a P(δi) ≤ 0.1 were removed, which closely translates to at least one system ranking a resource as first. The remaining resources were sorted in descending order.
Ontology collection and search queries
The PGT was compared against two sets of queries and ontologies. The first set included the same search queries and ontologies as the GT evaluation. The second was extended to include 51 extra queries obtained by [62] from the BioPortal query log (Table 6) and added 130 ontologies from the OBO Foundry (http://obofoundry.org/), which, considering the previous 23 ontologies, led to a collection of 153 ontologies.
Table 6.
Expanded query set obtained from [62]
| Type | Queries |
|---|---|
| General | Concentration unit, daily living, electron microscopy, health belief, health services, body weight, cell mass, cell proliferation, disease staging, dose response, clinical trial, compound treatment, differential scanning calorimetry, growth protocol, high-performance liquid, high throughput, sequence alignment |
| Cell or tissue | Bone marrow, brown adipose, connective tissue, connective tissue development, granulosa cell, haemoglobin E |
| Anatomy | Collecting duct, digestive system, embryonic structure, frontal lobe, harderian gland, heart ventricle |
| Genetic | Copy number, gene expression phenotype, gene regulation, genetic modification |
| Condition | Convulsive status epilepticus, fatty liver, generalized anxiety, heart failure, heart rate, venous thrombosis |
| Disease | Breast cancer, eye disease, haemoglobin E thalassaemia, hepatitis b, hepatitis c, ovarian cancer |
| Disorders | Cystathione synthase deficiency, Dowling-Degos syndrome, epileptic encephalopathy, fever infection syndrome, Goldstein-Hutt, nephrotic syndrome |
The method for building the PGT relies on the ability of systems to retrieve relevant results. Therefore, for this process, the search parameters were changed to return only exact matches.
Comparison between GT and PGT
The GT and the PGT were compared by calculating their Rank agreement and Relevancy agreement. These comparisons were performed over the collection of 10 queries and 23 ontologies. The evaluation results of both ground truths were compared using the Pearson correlation coefficient and the linear distance between the values obtained for each query with each algorithm/search application. The results were then averaged for each performance metric.
Results
The results are presented in relation to the GT, the comparison between the GT and the PGT and the extended results using only the PGT.
GT results
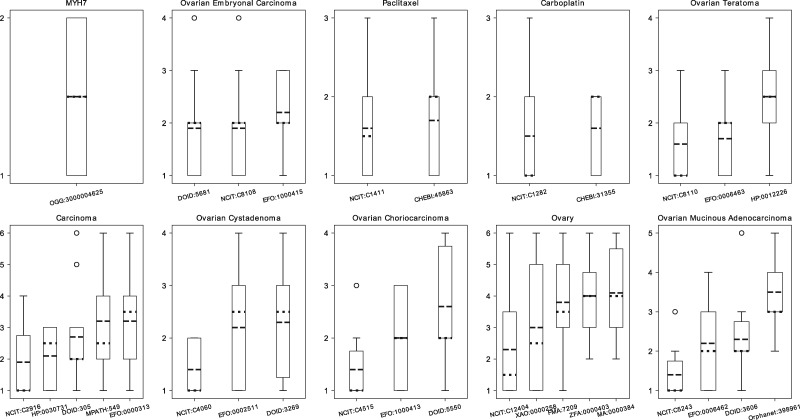
Figure 2 presents a box plot for each query with the respective answers that were included in the questionnaire. The results for each box plot are ordered by the mean, with ties not yet resolved. Overall, the queries show a high dispersion of answers. The query ‘MYH7’ had only one search result, and the opinions of the experts were divided equally between relevant and not-relevant. None of the means indicated that the judges, in average, considered a search result not relevant, i.e. the mean would have to be equivalent to the highest number in the options. The experts did not suggest more classes to be added to the preselected classes.
Figure 2.
Box plot of the results of the GT questionnaire. The y axis displays the possible number of ranks for each item (i.e number of answers plus the additional not-relevant rank). The x axis shows the class id for each of the possible answers for the queries. The dotted line represents the median, and the dashed line represents the mean by which the results were ordered.
Table 7 shows the rank obtained for the query ‘carcinoma’ by calculating the mean of the scores attributed to each class by the judges. If any mean calculation resulted in a draw, the final rank was obtained by searching the popularity of each ontology in BioPortal and ranking the most popular higher than the least popular. For example, the last two rankings in the Table 7 have a same mean value (3.2), and the EFO result was ranked above the MPATH because of EFO’s higher popularity in BioPortal.
Table 7.
Ranking of ‘Carcinoma’ in the GT
Validation
The observed agreement between the judges in relation to the rank was, in average, 30% with an SD of 20%. The relevancy agreement was, in average, 85%, with an SD of 15%. These results show that the judges have a high level of agreement when consider which classes are relevant or not-relevant but have a low agreement when ranking the ontology resources.
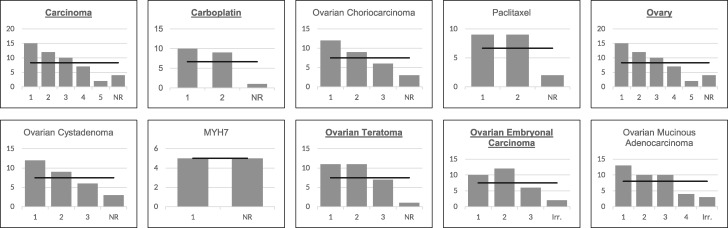
The expected values and the observed values of each query for the chi-square goodness-of-fit test are shown in Figure 3. The line represents the expected value, i.e. the value each rank should have if the rankings were equally chosen by the judges. The bars represent the actual number of times each rank was chosen. Except for the ‘MYH7’ query, the ranks in all queries differ from the expected value.
Figure 3.
Goodness-of-fit chi-square expected and observed results, represented by a line and bars, respectively. Each chart contains a bar for the number of rankings available for each query and one extra one representing the ranking of ‘Not-Relevant’ (NR). A bold and underlined query term indicates that the test rejected the null hypothesis, with α = 0.05.
The chi-square goodness-of-fit test indicated that, with α = 0.05, half of the answers reject the null hypothesis and the other half of the answers accept it, which implies that there is some disagreement between the judges. The ‘MYH7’ query had a χ2 of 0 because of the point previously raised of a single preselected relevant class. Queries with more general domains, such as ‘carcinoma’ and ‘ovary’, have a lower P-value, which strongly suggests that the judges’ responses are less likely to be random.
Comparison between GT and PGT
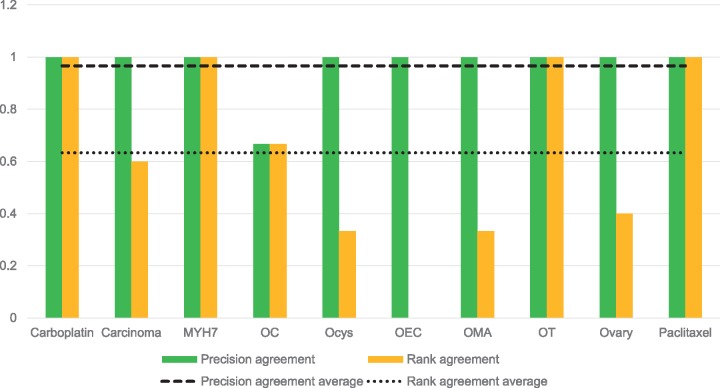
Figure 4 shows the relevancy and ranking agreement between the GT and the PGT for the collection. For all queries, except ‘Ovarian Choriocarcinoma’, the ground truths agree about which search results are considered relevant. ‘Ovarian Choriocarcinoma’ has a lower agreement because of the presence of the class ‘choriocarcinoma of ovary’ (http://purl.obolibrary.org/obo/DOID_5550), which is not an exact match. As the PGT was built based on exact matches only, this result was not considered and, therefore, is not featured on the final PGT. The rank agreement between ground truths is much lower than the relevancy agreement with an average of 63%.
Figure 4.
Relevancy and ranking agreement between the GT and the PGT.
NDCG uses the GT ranking to compute the iDCG. However, none of the remaining metrics takes into account the GT ranking to evaluate the performance of the algorithm/system. Therefore, because of the high relevancy agreement between GT and PGT, P@k, AP@k and MAP are considered more reliable than the NDCG when evaluating searches with the PGT.
Evaluation with performance metrics
Against the GT
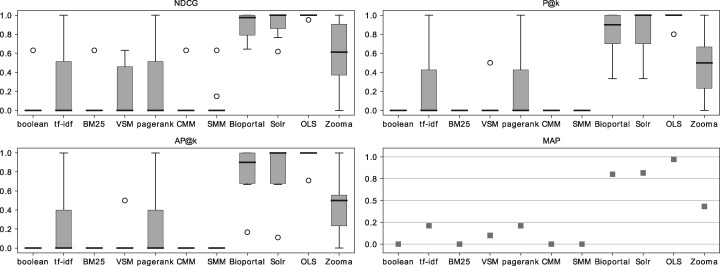
Tables 8 and 9 present the AP@3 and NDCG of each algorithm and application tested for the set of 10 queries. Figure 5 shows box plots for each of the metrics studied with consideration of partial and exact matches, and Figure 6 examines exact matches only.
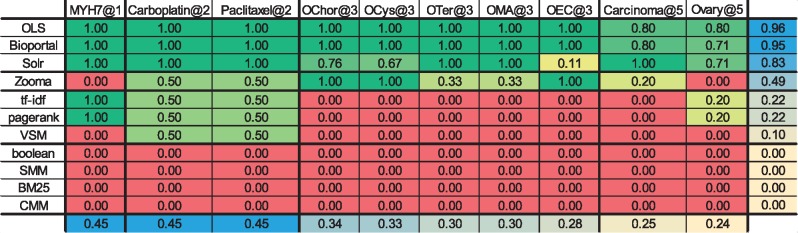
Table 8.
AP@3. The colours code the AP@3 values and range from dark green (highest AP@3, i.e. 1.0) to red (lowest AP@3, i.e. 0.0)
|
Note: The last column and last row represent the mean of each column/row, colour coded from blue (high mean) to light yellow (low mean).
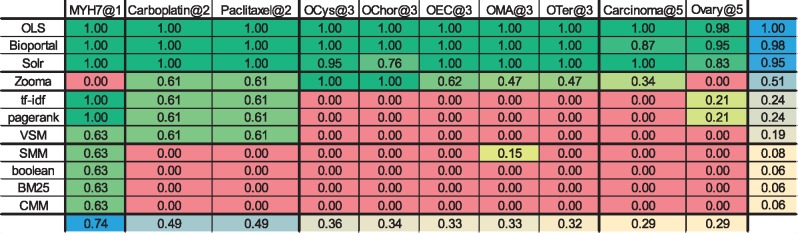
Table 9.
NDCG. The colours code the NDCG values and range from dark green (highest NDCG, i.e. 1.0) to red (lowest NDCG, i.e. 0.0)
|
Note: The last column and last row represent the mean of each column/row, colour coded from blue (high mean) to light yellow (low mean).
Figure 5.
NDCG, P@k, AP@k and MAP results for the 10 query collection, considering partial matches, against the GT.
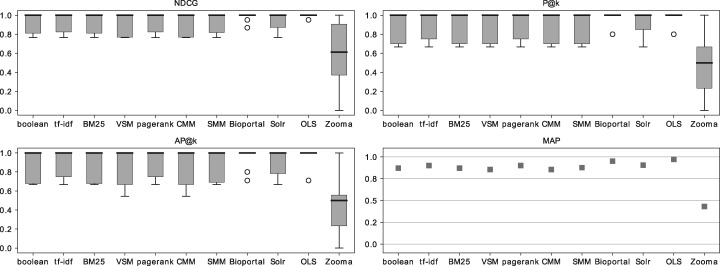
Figure 6.
NDCG, P@k, AP@k and MAP results for the 10 query collection, considering exact matches only, against the GT.
Algorithms results analysis
Tables 8 and 9 show that for the algorithms, most of the queries had an AP@3 and a NDCG equal to 0. The main contributors to these results were the partial matches. As none of these algorithms (except CMM) weights partial or full matches differently, a class that contained any of the query words was considered a match and was ranked according to the algorithm. Most of the algorithms, except tf-idf and VSM, also rank matches globally, which leads to several non-relevant results having a high score because of the global score of the ontology. CMM is one of the algorithms that rank ontologies globally, but as it evaluates the coverage of the set of queries by an ontology, it goes even further by considering the query set globally as well. ‘MYH7’, ‘Carboplatin’ and ‘Paclitaxel’ achieved the best AP@3 and NDCG results because of their low number of possible matches.
The comparison of Figure 5 with Figure 6 shows that forcing exact matches considerably increases the performance of the IR algorithms. However, the consequence of this change is that the algorithms and search applications ignore synonyms. By forcing exact matches, even the same label with a different order will not be returned by the algorithms. However, in the small scale considered, this was not a significant issue, as only one query matched with a synonym (http://www.ebi.ac.uk/efo/EFO_0002511— ‘simple cystadenoma’) and only one matched with a label with a different word order (http://purl.obolibrary.org/obo/DOID_5550— ‘choriocarcinoma of ovary’).
Search Applications Results Analysis
BioPortal focuses on precision, only showing the best hit in the ontologies with a match, while OLS focuses on recall, with the highest scoring terms ranked first, but also showing all possible partial matches after. These results mostly follow the same frequency pattern as Google’s results (Table 4), except for queries of type ‘Drug’, which have less or equal frequency as diseases. This can be explained by the number of query words in each search, as drugs only contain one word, the combinations of partial matches were limited. The search applications show high performance with MAP of 0.97 for OLS, 0.82 for Solr, 0.80 for Bioportal and 0.43 for Zooma.
NDCG evaluates the results considering not only the ontology classes present but also the position in which they appear. OLS achieved the highest NDCG performance with an average of 0.99 over all queries, BioPortal obtained an average NDCG of 0.90 and Solr with 0.92. Zooma obtained the lowest NDCG performance with an average of 0.58.
Figures 5 and 6 show that the difference between the results with or without forcing exact matches does not have a major effect in the search applications tested, as they were already ranking the exact and relevant matches in the first k positions. Zooma does not allow exact match-only search; therefore, the results are the same in both figures.
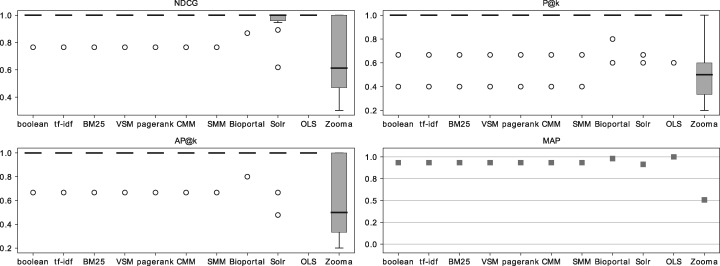
Against the PGT
Figure 7 presents the results for the collection of 10 query terms and 23 ontologies with forced exact matches against the PGT. Except for Zooma, the metrics show high performance for all algorithms and search applications. Overall, BioPortal and OLS slightly outperform the remaining methods with Zooma having the lowest performance in this setting.
Figure 7.
NDCG, P@k, AP@k and MAP results for the 10 query collection, considering exact matches only, against the PGT.
Figure 7 is directly comparable with Figure 6, as it used the same search parameters, but the results were compared with the PGT instead of the GT. Even though the box plots appear different, the medians are aligned and Table 10 shows a high correlation between the results with the GT and the PGT with a low average distance between the results. This correlation indicates that the results with the PGT show lower dispersion but are correlated to the results against the GT (which show higher dispersion in the box plot). The MAP comparison between the two figures also shows a high degree of similarity, with OLS and BioPortal slightly outperforming all other algorithms and Zooma obtaining the lowest results.
Table 10.
Correlation and average distance between GT and PGT results for each metric tested (P-value < 0.01)
| Metric | Pearson’s R | Average distance |
|---|---|---|
| NDCG | 0.75 | 0.03 |
| P@K | 0.64 | 0.07 |
| AP@K | 0.69 | 0.05 |
| MAP | 0.93 | 0.05 |
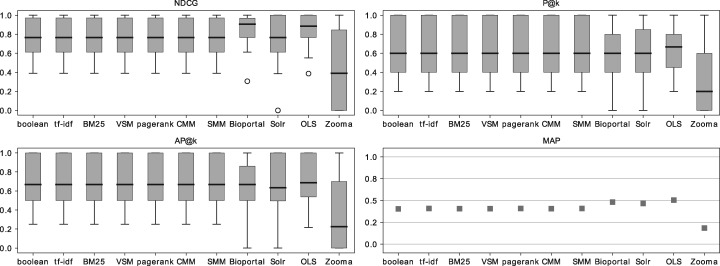
From these results we concluded that the PGT is a reliable complement for an expert-based GT in the setting described. Therefore, we extended the number of queries and ontology collection and tested the algorithms and search applications against the respective PGT. Figure 8 shows that the overall conclusions of the extended search are similar to the ones obtained with the smaller query set. The search applications achieve a superior performance against the IR algorithms, with OLS having the best performance, followed by BioPortal, Solr and Zooma having the lowest results.
Figure 8.
NDCG, P@k, AP@k and MAP results for the extended query and ontology collection, considering exact matches only, against the PGT.
Discussion
These are some of the key search factors which influenced the process of ranking resources in ontologies.
Ground truth
Some judges ranked a class as not relevant, while other judges chose the same class as the most relevant. The principles of ontology engineering guide that ontologies should be orthogonal, which means that the same concept should not be independently created in each ontology, but, if it already exists, it should be reused from existing ontologies. In reality, however, ontologies are neither complete nor orthogonal, and labels do not have a perfect representation of the semantics of a concept. The exact same concept can, therefore, be described in different ontologies, and experts do not agree, which one should be the unifying one, as some experts will have different interpretations of the semantics of the label or the semantic categorization of the ontology of a concept. This disagreement makes the task of building a gold standard for ontology search results tough.
Probabilistic ground truth
The main goal of building a PGT was to perform a deeper evaluation without having more human experts. Finding experts both in the biomedical domain and knowledge engineering to fill an extensive questionnaire is a non-trivial task. The PGT was shown to have a significant agreement with the GT, and the extended search showed that even with a broader domain of queries, the search applications still outperform the IR algorithms, and OLS and BioPortal are the best performing systems.
Partial matches
The performance of all the IR algorithms suffered from too many partial matches. In biomedical ontologies, it is common to find multi-word labels, as this domain describes complex concepts such as different phenotypes or anatomical parts. For example, when the input for the algorithms is the query ‘Ovarian Cystadenoma’, the results consist mostly of partial matches of the word ‘ovarian’. The large number of partial matches led to a precision and NDCG of 0 for most algorithms tested. The CMM algorithm is the only one in this set that distinguishes between partial and exact matches. However, it did not perform better than others, as it does not evaluate each query individually. The relevance of partial matches to the performance of the IR algorithms was demonstrated with the limitation of the search to exact matches only. However, this performance change cannot be considered usable, as search through exact matches only exclude the complex semantics (i.e. synonyms or descriptions) of ontologies from the search.
Tie-breaking
In the GT, ties were resolved by ordering them by ontology popularity, which was obtained from BioPortal. This method is also the main boost factor for search results in BioPortal. In this context, this method did not create a bias towards BioPortal, as the number of ties was low (only two ties for all 10 query words) and the remaining search applications also had a good overall performance, with OLS slightly outperforming BioPortal.
Controlled versus open-access ontologies
The search process included a set of 23 ontologies that was later expanded to 153 open-access ontologies. BioPortal contains several restricted access ontologies (e.g. SNOMED CT) that were featured in the search results but could not be included in the GT and, therefore, could not be evaluated. In some cases, this means that even though the GT contains some of the possible classes, BioPortal can have several more, and there was no way, at this point, to evaluate their relevance in relation to the open-access ontologies.
General versus specialized terminologies
The search applications tested did not agree on the ranking for some queries. This issue was more noticeable when the query was more general, such as in the search for ‘ovary’. This query has several exact matches in different ontologies, and all of the applications ranked them differently. Some of those matches are species-specific, but their descriptions are general. Table 11 shows the comparison of the results of the distinct applications tested, the GT and the PGT.
Table 11.
Comparing the ranking for ‘ovary’ between the GT, BioPortal (BP), OLS, Solr and PGT
| Class URI | GT | BP | OLS | Solr | PGT |
|---|---|---|---|---|---|
| http://purl.obolibrary.org/obo/NCIT_ C12404 | 1 | 1 | 1 | 3 | 1 |
| http://purl.obolibrary.org/obo/XAO_ 0000258 | 2 | 4 | 3 | 2 | 4 |
| http://purl.obolibrary.org/obo/FMA_ 7209 | 3 | 2 | 5 | - | 5 |
| http://purl.obolibrary.org/obo/ZFA_ 0000403 | 4 | 3 | 4 | 1 | 3 |
| http://purl.obolibrary.org/obo/MA_ 0000384 | 5 | - | 2 | 4 | 2 |
Solr considerations
Solr achieved a good performance, and its biggest advantage is that any user can index and search through their own set of ontologies.
OLS considerations
OLS uses Solr as its base for indexing and searching and boosts specific ontologies. The value of the boost is unknown, and the process of attributing a boost to an ontology is not explicitly explained.
Zooma considerations
Zooma’s performance suffered not only from obtaining matches from annotated data but also from its focus on high precision. For all queries that found a match in the curated data, the search returned only one or two results. In most cases, these results did not match what the experts considered to be the most relevant ontology class. With only eight data sources to choose from, it is possible that the annotated data focused on domains not represented by the queries used. The queries that achieve top scores with Zooma are the queries that did not match any term in the curated data and, therefore, Zooma used OLS to find matches for the query.
Conclusion and recommendations
The article evaluates and reviews seven state-of-the-art ranking algorithms and four search applications. The article established a GT through a user study with 10 experts that ranked 10 cancer-related queries and also established a GT based on the consensus from the algorithms and systems tested. The ground truths were compared against the results from the noranking algorithms and the search applications. The evaluation experiment used 61 search queries (10 terms from the cancer genomics domain plus 51 general biomedical terms) and 153 biomedical ontologies (23 ontologies related to cancer genomics terms plus 130 general biomedical ontologies). Based on this analysis, we are able to conclude that (1) in their current state, the algorithms cannot handle partial matches, but forcing exact matches boosts their performance with possible loss of information, and (2) the search applications are already robust in finding the relevant concepts for search queries in the correct order, with high precision and recall. The performance of search applications severely degrades with ambiguous search queries when compared with specific/concise queries. After evaluating the technologies, we conclude that, even though BioPortal and OLS outperform all other applications, one should not be chosen over the others by performance alone, but instead each situation (i.e. search scenario) should be analysed to choose which application to use:
Searching for top-K
Both BioPortal and OLS have good precision in the top three hits, but both of them return a lot more results for general queries. It is possible to tune both tools to only return exact matches to reduce the number of matches, but the applications can still obtain more classes than the user is expecting. On the other hand, Zooma’s smaller repository returns only one or two results, but, that means that the queries have to be tuned towards the domains annotated by the curated data.
Set of ontologies
If the set of ontologies used is a restriction for the search, BioPortal, OLS and Zooma can filter the ontologies shown in the results. Besides the ontologies available in OLS, Zooma also includes data sources used for the annotation process. OLS, however, does not index part of the ontologies indexed by BioPortal because of (1) BioPortal allowing user-submitted ontologies and OLS curating the ontologies allowed in the system, and (2) BioPortal having a large set of relevant licenced ontologies (e.g. SNOMED CT), which are not indexed by OLS and, unless the user has access, cannot be indexed with Solr. Both of these conditions should be taken into account when choosing which service to use. If the user wants to use only open-access ontologies, it can filter them in BioPortal, but more easily can just search through all the indexed ontologies in OLS. However, if the user wants to search the largest possible set of ontologies, BioPortal would be the suggested choice.
Looking for partial matches
When the main goal of the search is not to find an exact match but to find related terms to the one being searched, OLS is the best solution. Contrary to BioPortal, which shows only the most relevant class in each ontology, OLS ranks and shows every possible match within the ontologies it has indexed. High-scoring matches are shown first and then every possible partial match is also displayed.
Custom set of ontologies
In a hypothetical scenario where a user has a custom ontology, Solr would be the most appropriate choice. Solr allows the users to index their ontologies with no restrictions.
Search with curated annotations
Despite Zooma’s lower performance in this context, it can be applied in a situation where a user would rather have a previously curated annotation to map to a term than just a search result generated by an algorithm. When possible, Zooma returns mappings that match the term and that have been previously annotated in different repositories.
Future work
In the future, to consolidate the GT, we will study other methods of breaking ties. One possibility is to apply the CMM algorithm to get the coverage of the ontology for a set of queries. The ontology with the highest CMM score would be ranked higher. Another option could be to search the LOD Cloud for popularity of the ontology classes to rank them according to real-world usage. Further algorithms and techniques could be studied to infer how the popularity-based tie-breaking affects the results. A framework integrated with an application, such as Solr or ElasticSearch, could also be useful to streamline the process of indexing and searching through a customized set of ontologies and control the search process. Furthermore, testing with the ranking algorithms could also lead to the development of a standard technique to rank some of the more generic queries to obtain a uniform ranking among the different search applications.
We believe that the proposed benchmark is a good indicator of the performance of biomedical ontology search applications, and we expect that this evaluation will allow researchers to apply the current search techniques more reliably and that finding the best application to fit their annotation needs will be easier. We hope that with this work ontology engineers will also be better informed on how to find and use ontological resources in their research work.
Key Points.
The article is the first attempt to evaluate four ontology search applications and seven IR algorithms on their ability to retrieve top-ranked search results.
A total of 61 search queries and 153 biomedical ontologies are evaluated against four ontology search applications and seven IR algorithms.
An extensive judgement based on the opinions of experts and automatically generated GT is derived evaluating the overall performance of ontology search applications and IR algorithms.
The set of ontologies available is an important criterion for selection the system. If the user requires the largest set of ontologies, they should choose Bioportal. If they require every possible exact and partial match, OLS is the most appropriate. Solr would be the most appropriate when the user has a custom set of ontologies.
The article identifies key search factors for biomedical ontologies that will help biomedical experts and ontology engineers to select the best-suited search application and/or algorithm in different search scenarios.
Daniela Oliveira is a PhD Student at the Insight Centre for Data Analytics, Galway. Daniela has a background in health sciences and bioinformatics and is currently working in the field of biomedical ontologies.
Anila Sahar Butt received the PhD degree from the Australian National University. She is currently a Postdoctoral Fellow at CSIRO, Australia. Her research interests include ontology search and ranking.
Armin Haller is an MBA Director and a Senior Lecturer at the Australian National University with a joint appointment at the Research School of Management and the Research School of Computer Science.
Dietrich Rebholz-Schuhmann is a Medical Doctor and a Computer Scientist. Currently, he is established chair for data analytics at the National University of Ireland, Galway, and the director of the Insight Center for Data Analytics in Galway. His research is positioned in semantic technologies in the biomedical domain. In his previous research, he has established large-scale on-the-fly biomedical text mining solutions and has contributed to the semantic normalization in the biomedical domain.
Ratnesh Sahay is the Head of research unit, Research fellow and Adjunct lecturer at the Insight Centre for Data Analytics, National University of Ireland, Galway. He is the lead scientist of BIOOPENER project (http://bioopenerproject.insight-centre.org/), and his research focuses on using semantics for solving key data integration, interoperability and analytics challenges in the ehealth, clinical trial and biomedical domains.
Funding
This work has been supported by the Science Foundation Ireland (grant number SFI/12/RC/2289).
References
- 1. Smith B, Ceusters W, Klagges B, et al. . Relations in biomedical ontologies. Genome Biol 2005;6(5):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Szolovits P. Artificial Intelligence in Medicine. Boulder, CO: Westview Press, 1982. [Google Scholar]
- 3. Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform 2008;67–9. [PMC free article] [PubMed] [Google Scholar]
- 4. Ashburner M, Ball CA, Blake JA, et al. . Gene Ontology: tool for the unification of biology. Nat Genet 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoehndorf R, Schofield PN, Gkoutos GV.. The role of ontologies in biological and biomedical research: a functional perspective. Brief Bioinform 2015;16(6):1069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marshall MS, Boyce RD, Deus HF, et al. . Emerging practices for mapping and linking life sciences data using RDF—a case series. Web Semant 2012;14:2–13. [Google Scholar]
- 7. Hu W, Qiu H, Dumontier M. Link analysis of life science linked data. In: Proceedings of The Semantic Web - ISWC 2015, Bethlehem, PA, USA, October 11-15, 2015, vol. 9367 Springer, 2015, 446–62.
- 8. Butt AS, Haller A, Xie L. Ontology search: an empirical evaluation. In: The Semantic Web - ISWC 2014, Riva del Garda, Italy, October 19-23, 2014. Volume 8797 of Lecture Notes in Computer Science Springer, 2014, 130–47.
- 9. Zaragoza H, Rode H, Mika P, et al. Ranking very many typed entities on wikipedia. In: Proceedings of the Sixteenth ACM Conference on Conference on Information and Knowledge Management. ACM, Lisbon, Portugal, 2007, 1015–18.
- 10. Ding L, Finin T, Joshi A, et al. Swoogle: a search and metadata engine for the semantic web. In: Proceedings of the 13th ACM International Conference on Information and Knowledge Management ACM, New York, NY, 2004, 652–9.
- 11. Tummarello G, Delbru R, Oren E. Sindice.Com: weaving the open linked data. In: Proceedings of the 6th International the Semantic Web Conference Springer-Verlag, Berlin, Heidelberg, 2007, 552–65.
- 12. d’Aquin M, Motta E.. Watson, more than a semantic web search engine. Semantic Web 2011;2(1):55–63. [Google Scholar]
- 13. Harth A, Umbrich J, Hogan A, et al. Yars2: a federated repository for querying graph structured data from the web. In: The Semantic Web: 6th International Semantic Web Conference, 2nd Asian Semantic Web Conference, ISWC 2007 + ASWC 2007, Busan, Korea, November 11-15, 2007, Proceedings. Springer-Verlag, Berlin, Heidelberg, 211–24.
- 14. Page L, Brin S, Motwani R, et al. The PageRank citation ranking: bringing order to the web. Technical report, Stanford InfoLab, 1999.
- 15. Qu Y, Cheng G.. Falcons concept search: a practical search engine for web ontologies. IEEE Trans Syst Man Cybern A Syst Hum 2011;41(4):810–6. [Google Scholar]
- 16. Gangemi A, Catenacci C, Ciaramita M, et al. A theoretical framework for ontology evaluation and validation. In: Proceedings of the 2nd Italian Semantic Web Workshop, Trento, Italy, volume 166 of CEUR Workshop Proceedings CEUR-WS.org, 2005.
- 17. Guarino N, Welty C.. Evaluating ontological decisions with OntoClean. Commun ACM 2002;45(2):61–5. [Google Scholar]
- 18. Lozano-Tello A, Gomez-Perez A.. ONTOMETRIC: a method to choose the appropriate ontology. J Database Manag 2004;15(2):1–18. [Google Scholar]
- 19. Alani H, Brewster C, Shadbolt N.. Ranking ontologies with AKTiveRank In: The Semantic Web - ISWC 2006. Berlin, Heidelberg: Springer, 2006, 1–15. [Google Scholar]
- 20. Suchanek FM, Kasneci G, Weikum G. Yago: a core of semantic knowledge. In: Proceedings of the 16th International Conference on World Wide Web, WWW’07 ACM, New York, NY, 2007, 697–706.
- 21. Patel C, Supekar K, Lee Y, et al. Ontokhoj: a semantic web portal for ontology searching, ranking and classification. In: Proceedings of the 5th ACM International Workshop on Web Information and Data Management. ACM, New Orleans, Louisiana, USA, 2003, 58–61.
- 22. Thomas E, Pan JZ, Sleeman D. ONTOSEARCH2: Searching Ontologies Semantically. In: Proceedings of the OWLED 2007 Workshop on OWL: Experiences and Directions, Innsbruck, Austria, 2007. [Google Scholar]
- 23. Buitelaar P, Eigner T, Declerck T. Ontoselect: A dynamic ontology library with support for ontology selection. In: Proceedings of the Demo Session at the International Semantic Web Conference, Hiroshima, Japan, 2004.
- 24. Buitelaar P, Eigner T. Evaluating ontology search. In: Proceedings of the 5th International Workshop on Evaluation of Ontologies and Ontology-Based Tools, Busan, Korea, 2007, 11–20.
- 25. Whetzel PL, Noy NF, Shah NH, et al. . BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res 2011;39(suppl):W541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jupp S, Burdett T, Leroy C, et al. A new ontology lookup service at EMBL-EBI. In: Proceedings of SWAT4LS International Conference 2015 CEUR-WS.org, Cambridge, UK, 2015, 118–19.
- 27. Adamusiak T, Burdett T, Kurbatova N, et al. . OntoCAT–simple ontology search and integration in Java, R and REST/JavaScript. BMC Bioinformatics 2011;12(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kurbatova N, Adamusiak T, Kurnosov P, et al. . ontocat: an R package for ontology traversal and search. Bioinformatics 2011;27(17):2468–70. [DOI] [PubMed] [Google Scholar]
- 29. Petryszak R, Burdett T, Fiorelli B, et al. . Expression atlas update-a database of gene and transcript expression from microarray-and sequencing-based functional genomics experiments. Nucleic Acids Res 2014;42(D1):D926–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MacArthur J, Bowler E, Cerezo M, et al. . The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog). Nucleic Acids Res 2017;45(D1):D896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salton G, Buckley C.. Term-weighting approaches in automatic text retrieval. Inf Process Manag 1988;24(5):513–23. [Google Scholar]
- 32. Robertson S, Walker S, Jones S, et al. Okapi at TREC-3, January 1995. https://www.microsoft.com/en-us/research/publication/okapi-at-trec-3/.
- 33. Salton G, Wong A, Yang CS.. A vector space model for automatic indexing. Commun ACM 1975;18(11):613–20. [Google Scholar]
- 34. Page L, Brin S, Motwani R, et al. The PageRank Citation Ranking: Bringing Order to the Web, November 1999. http://ilpubs.stanford.edu: 8090/422/.
- 35. Courtot M, Gibson F, Lister AL, et al. . MIREOT: the minimum information to reference an external ontology term. Appl Ontol 2011;6(1):23–33. [Google Scholar]
- 36. Horridge M, Bechhofer S.. The owl api: a java API for owl ontologies. Semant Web 2011;2(1):11–21. [Google Scholar]
- 37. Hastings J, de Matos P, Dekker A, et al. . The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res 2013;41(D1):D456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bard J, Rhee SY, Ashburner M.. An ontology for cell types. Genome Biol 2005;6(2):R21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schriml LM, Arze C, Nadendla S, et al. . Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res 2012;40(D1):D940–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hanna J, Joseph E, Brochhausen M, et al. . Building a drug ontology based on rxnorm and other sources. J Biomed Semant 2013;4(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ison J, Kalaš M, Jonassen I, et al. . EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics 2013;29(10):1325–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Malone J, Holloway E, Adamusiak T, et al. . Modeling sample variables with an experimental factor ontology. Bioinformatics 2010;26(8):1112–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosse C, Mejino JL.. A reference ontology for biomedical informatics: the foundational model of anatomy. J Biomed Inform 2003;36(6):478–500. [DOI] [PubMed] [Google Scholar]
- 44. Köhler S, Doelken SC, Mungall CJ, et al. . The human phenotype ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 2013;42(D1):D966–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hayamizu TF, Baldock RA, Ringwald M.. Mouse anatomy ontologies: enhancements and tools for exploring and integrating biomedical data. Mamm Genome 2015;26(9–10):422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith CL, Goldsmith CA, Eppig JT.. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol 2004;6(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schofield PN, Sundberg JP, Sundberg BA, et al. . The mouse pathology ontology, MPATH; structure and applications. J Biomed Semant 2013;4(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gkoutos GV, Schofield PN, Hoehndorf R.. The neurobehavior ontology: an ontology for annotation and integration of behavior and behavioral phenotypes. Int Rev Neurobiol 2012;103:69–87. [DOI] [PubMed] [Google Scholar]
- 49. Sioutos N, de Coronado S, Haber MW, et al. . NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information. J Biomed Inform 2007;40(1):30–43. [DOI] [PubMed] [Google Scholar]
- 50. He Y, Sarntivijai S, Lin Y, et al. . OAE: the ontology of adverse events. J Biomed Semant 2014;5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He Y, Liu Y, Zhao B. OGG: a biological ontology for representing genes and genomes in specific organisms. In: Proceedings of the 5th International Conference on BiomedicalOntologies (ICBO), CEUR Workshop Proceedings, Houston, TX, USA, 2014, 13–20.
- 52. Mungall CJ, Gkoutos GV, Smith CL, et al. . Integrating phenotype ontologies across multiple species. Genome Biol 2010;11(1):R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Avraham S, Tung CW, Ilic K, et al. . The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res 2008;36:D449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Haendel MA, Gkoutos GG, Lewis SE, Mungall C Uberon: towards a comprehensive multi-species anatomy ontology. In: International Consortium of Biomedical Ontology, Nature Proceedings, Buffalo, New York, 2009. [Google Scholar]
- 55. Park CA, Bello SM, Smith CL, et al. . The Vertebrate Trait Ontology: a controlled vocabulary for the annotation of trait data across species. J Biomed Semant 2013;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schindelman G, Fernandes JS, Bastiani CA, et al. . Worm Phenotype Ontology: integrating phenotype data within and beyond the C. elegans community. BMC Bioinformatics 2011;12(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Segerdell E, Bowes JB, Pollet N, et al. . An ontology for Xenopus anatomy and development. BMC Dev Biol 2008;8:92. ISSN 1471-213X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Van Slyke CE, Bradford YM, Westerfield M, et al. . The zebrafish anatomy and stage ontologies: representing the anatomy and development of Danio rerio. J Biomed Semant 2014;5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim JH. Chi-square goodness-of-fit tests for randomly censored data. Ann Stat 1993;21(3):1621–39. [Google Scholar]
- 60. Lamiroy B, Sun T.. Computing precision and recall with missing or uncertain ground truth In: Graphics Recognition. New Trends and Challenges. Springer-Verlag, Berlin, Heidelberg, 2013, 149–62. [Google Scholar]
- 61. Wang Y, Wang L, Li Y, et al. A theoretical analysis of ndcg ranking measures. In: Proceedings of the 26th Annual Conference on Learning Theory (COLT 2013), Princeton, NJ, USA, 2013.
- 62. Gavankar C, Li YF, Ramakrishnan G. Explicit query interpretation and diversification for context-driven concept search across ontologies. In: Groth P. et al. (eds) The Semantic Web - ISWC 2016. ISWC 2016. Lecture Notes in Computer Science, Vol. 9981. Springer, Cham, 2016, 271–88.