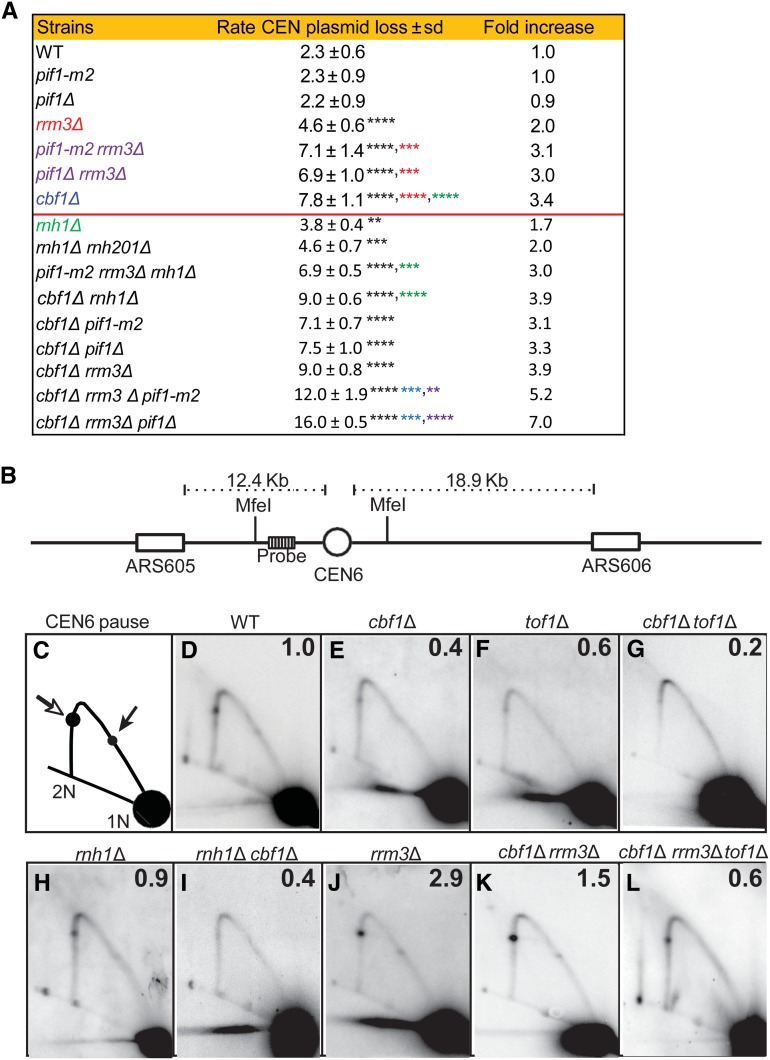
Figure 5.
Stability of a CEN6 plasmid and 2D gel analysis of replication of the chromosomal CEN6 locus, in mutant and WT strains. Methods for determining CEN6 plasmid loss rates and 2D gels are described in the Materials and Methods, and in Chen et al. (2019). (A) CEN plasmid loss rate in WT and mutant strains growing at 30° in YPD. Loss rates for strains above the horizontal red line are from Chen et al. (2019). The second column is the loss rate ± 1 SD for the indicated strain. Asterisks indicate significant differences: black asterisks are significant differences in loss rate of the indicated strain compared to loss rate in WT cells; red asterisks are significant differences between indicated strain and rrm3Δ cells; green asterisks are significant differences between indicated strain and rnh1Δ cells; blue asterisks are significant differences between indicated strain and cbf1Δ cells; and purple asterisks indicate significant differences between indicated strain and pif1Δ (or pif1-m2) rrm3Δ cells. (B) 2D gel electrophoresis and Southern blot hybridization of replication through the chromosomal CEN6 locus in 30°-grown asynchronous WT and mutant cells growing in YPD. DNA was isolated, digested with MfeI restriction enzyme, and analyzed by 2D gels and Southern blotting. (B) Schematic of the chromosomal MfeI fragment that contains CEN6 (open circle) relative to the replication origins (open boxes) on either side. The position of the radiolabeled probe used for Southern blot analysis is indicated. (C) Schematic of 2D gel signal of MfeI-digested DNA using the probe indicated in (B). Arrows mark the pauses at CEN6 along the arc of Y-shaped replication intermediates. The pause produced from replication forks originating from ARS605 are indicated by the open arrow, whereas the pause arising from forks originating from ARS606 are indicated by the solid arrow. 1N indicates nonreplicating linear MfeI fragments. 2N indicates near-fully replicated MfeI fragments. (D–K) Southern blots of 2D gels from cells of the following genotypes: (D) WT, (E) cbf1Δ, (F) tof1Δ, (G) cbf1Δ tof1Δ, (H) rnh1Δ, (I) rnh1Δ cbf1Δ, (J) rrm3Δ, (K) cbf1Δ rrm3Δ, and (L) cbf1Δ rrm3Δ tof1Δ. The signal at the pause was quantified (Tran et al. 2017) and normalized to the pause signal in WT cells to obtain the relative fold change. The average fold difference of mutant over WT from two or more independent biological replicates is shown in the upper right corner of each panel. 2D gel data for tof1Δ and rrm3Δ were published previously (Chen et al. 2019). 2D, two-dimensional; WT, wild-type.