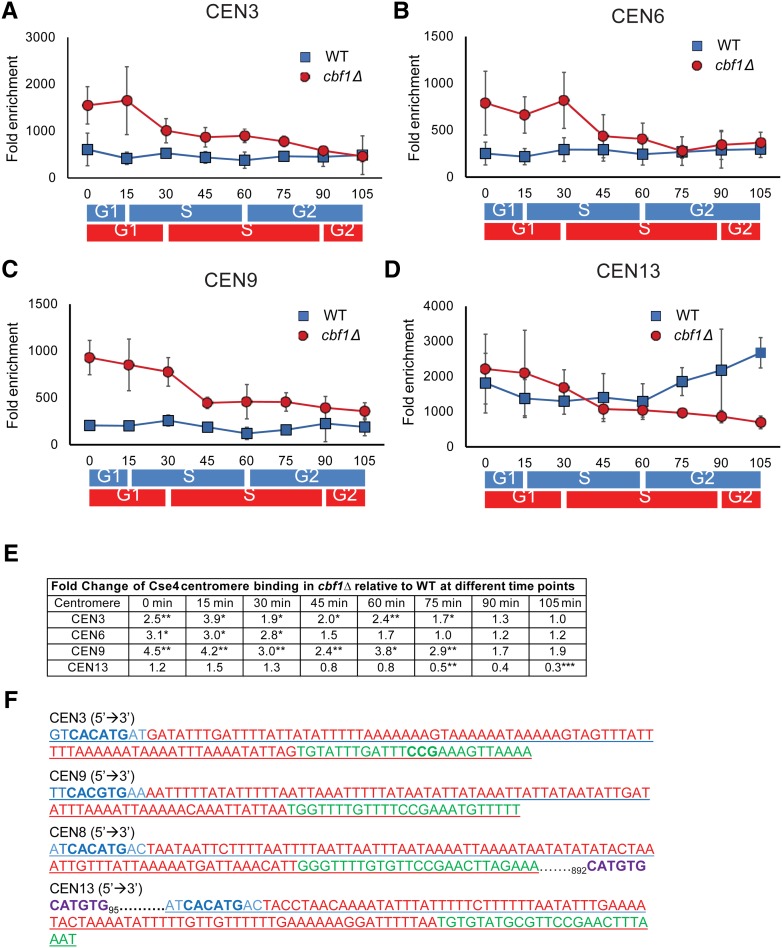
Figure 7.
The timing of Cse4 binding is altered in synchronized cbf1Δ cells. Myc9-tagged Cse4 binding to different centromeres was determined by ChIP in synchronized cells grown at 24° in YPD in WT and cbf1Δ cells. Data are presented as ([ChIP/Input]Target site/[ChIP/Input]ARO). The binding of Cse4-Myc9 in WT (blue squares) and cbf1Δ (red circles) cells at (A) CEN3, (B) CEN6, (C) CEN9, and (D) CEN13. Blue and red bars indicate approximate positions in the cell cycle as determined by FACS in, respectively, WT and cbf1Δ cells (see Figure S1A). For CEN3 (A) and CEN9 (C), binding was significantly higher in cbf1Δ cells than in WT cells from 0 to 75 min (P < 0.05). For CEN6 (B), binding was significantly higher in cbf1Δ cells than in WT cells from 0 to 30 or 45 min (P < 0.05). At CEN 13 (D), Cse4 binding was significantly lower in cbf1Δ cells than in WT cells only at 75 and 105 min (P < 0.01). However, binding of Cse4 to CEN13 in WT cells was significantly higher than to any of the three other centromeres throughout the cell cycle (P < 0.05). (E) Table showing time points for each centromere where Cse4 binding was significantly different from that in WT cells. For each time point, fold change is the difference at that time point in cbf1Δ cells compared to WT. (F) Analysis of sequences for CEN3 (chosen because it has a nonpalindromic CDEI site), CEN9 (chosen because it has a palindromic CDEI site), CEN8, and CEN13. Blue letters indicate CDEI site. Red color indicates CDEII site. Green color indicates CDEIII site. Purple color indicates a second potential Cbf1-binding site. Other than CEN13, none of the other 15 centromeres has an additional CDEI consensus site < 1000 bp from the core centromere. CDE, conserved elements; ChIP, chromatin immunoprecipitation; WT, wild-type.