Structural maintenance of chromosomes flexible hinge domain-containing l (SMCHDl) is an architectural factor critical for X chromosome inactivation (XCI). In mice, loss of all Smchdl causes female-specific embryonic lethality due to an XCI defect. However...
Keywords: SMCHD1, X-chromosome inactivation, HOX genes, clustered protocadherin genes, epigenetics, chromatin
Abstract
Structural maintenance of chromosomes flexible hinge domain-containing 1 (SMCHD1) is an architectural factor critical for X-chromosome inactivation (XCI) and the repression of select autosomal gene clusters. In mice, homozygous nonsense mutations in Smchd1 cause female-specific embryonic lethality due to an XCI defect. However, although human mutations in SMCHD1 are associated with congenital arhinia and facioscapulohumeral muscular dystrophy type 2 (FSHD2), the diseases do not show a sex-specific bias, despite the essential nature of XCI in humans. To investigate whether there is a dosage imbalance for the sex chromosomes, we here analyze transcriptomic data from arhinia and FSHD2 patient blood and muscle cells. We find that X-linked dosage compensation is maintained in these patients. In mice, SMCHD1 controls not only protocadherin (Pcdh) gene clusters, but also Hox genes critical for craniofacial development. Ablating Smchd1 results in aberrant expression of these genes, coinciding with altered chromatin states and three-dimensional (3D) topological organization. In a subset of FSHD2 and arhinia patients, we also found dysregulation of clustered PCDH, but not HOX genes. Overall, our study demonstrates preservation of XCI in arhinia and FSHD2, and implicates SMCHD1 in the regulation of the 3D organization of select autosomal gene clusters.
X-CHROMOSOME inactivation (XCI) silences gene expression on one of the two X chromosomes in female mammals, thereby ensuring balanced dosage of X-linked genes between XY males and XX females (Starmer and Magnuson 2009; Lee 2011; Wutz 2011; Disteche 2012; Mira-Bontenbal and Gribnau 2016). Aberrant dosage of even a fraction of X-linked genes in humans can produce an abnormal phenotype, as exemplified by Turner (XO) and Klinefelter (XXY) syndromes. In mice, perturbation of XCI in early development has devastating consequences: deletion of Xist (X-inactive-specific transcript), the master regulator of XCI (Brockdorff et al. 1991; Brown et al. 1991), causes female-specific embryonic lethality early in gestation around the time of normal XCI initiation (Marahrens et al. 1997; Yang et al. 2016).
A mutagenesis screen in mice previously identified Smchd1 as a locus essential for transgene silencing and female embryonic development (Blewitt et al. 2005, 2008). Smchd1 encodes structural maintenance of chromosomes (SMC) flexible hinge domain-containing 1 (SMCHD1), a 230-kDa protein with an N-terminal GHKL (Gyras, Hsp90, Histidine Kinase, MutL)-type ATPase domain and a C-terminal SMC hinge domain that mediates SMCHD1 homodimerization (Blewitt et al. 2008; Brideau et al. 2015; Chen et al. 2015, 2016). Several studies have now demonstrated its unequivocal role in XCI (Blewitt et al. 2005, 2008; Gendrel et al. 2012, 2013; Mould et al. 2013; Nozawa et al. 2013; Jansz et al. 2018a,b; Sakakibara et al. 2018; Wang et al. 2018; Gdula et al. 2019). Indeed, SMCHD1 is enriched on the inactive X chromosome (Xi) (Blewitt et al. 2008), but is apparently recruited to the Xi relatively late during XCI in an ex vivo female embryonic stem (ES) cell model (Gendrel et al. 2012). In contrast to Xist mutants, female mice null for Smchd1 do not perish during the peri-implantation stage, but rather only in midgestation [by embryonic day (E)11.5] (Blewitt et al. 2005, 2008). Thus, SMCHD1 is thought to act later during the XCI cascade. Indeed, recent work has shown that SMCHD1 functions in finalizing the three-dimensional (3D) reorganization the Xi, in particular at the level of chromosome compartments (Wang et al. 2018, 2019). During XCI, Xist recruits Polycomb Repressive Complex 1 to fuse A/B compartments into “S1/S2” compartments. SMCHD1 then binds S1/S2 structures and merges them to create a compartment-less Xi architecture. Loss of SMCHD1 and a failure to fuse S1/S2 leads to regional defects in Xist spreading and erosion of heterochromatic silencing, coinciding with derepression of 43% of Xi genes in SMCHD1-deficient cells (Wang et al. 2018). Thus, SMCHD1 plays a critical role during the progression through XCI.
Importantly, SMCHD1 shows a high degree of sequence similarity between mice and humans (87% protein sequence identity). SMCHD1 also associates with the Xi in both species (Blewitt et al. 2008; Nozawa et al. 2013). Given the conservation of structure and function across species, SMCHD1 mutations in humans would be expected to cause defective XCI and possibly female-specific phenotypes. A review of the human Genome Aggregation Database (gnomAD) of nearly 140,000 control exomes and whole genomes reveals that SMCHD1 is strongly depleted for loss-of-function (LoF) mutations (probability of being loss of function intolerant (pLI) = 1; observed/expected constraint metric (o/e) = 0.09), and that there is complete absence of individuals with homozygous LoF variants. These data suggest that a complete loss of SMCHD1 activity may be incompatible with life in either sex.
Human genetic studies over the last few years have now established a diverse phenotypic spectrum of complex disorders associated with SMCHD1 mutations. It was previously established that missense and LoF mutations in SMCHD1 are a necessary but insufficient component of a complex oligogenic mechanism associated with facioscapulohumeral muscular dystrophy type 2 (FSHD2) (Lemmers et al. 2012; Sacconi et al. 2013), a late-onset, slowly progressive muscle disease. SMCHD1 mutations also serve as a genetic modifier of disease severity in FSHD type 1 (Sacconi et al. 2013). Surprisingly, it has been recently demonstrated that some of these same FSHD2 mutations, and other missense variants of SMCHD1, contribute to a rare craniofacial malformation that involves the complete absence of an external nose, known as congenital arhinia (Gordon et al. 2017; Shaw et al. 2017). While FSHD2 and arhinia-associated mutations are only found in the heterozygous state in humans, heterozygous mutations can be equally or more detrimental than null mutations, either through haploinsufficiency or a dominant-negative mechanism (for instance, interference with SMCHD1 homodimerization) (Wilkie 2017). Indeed, several studies suggest that the phenotypes associated with arhinia and FSHD2 likely arise from either a haploinsufficiency or a dominant-negative effect (Lemmers et al. 2012, 2015; Shaw et al. 2017; de Greef et al. 2018).
An intriguing aspect of these disorders is that none exhibits a sex bias, despite the established role of SMCHD1 in XCI. Could SMCHD1 dysfunction cause severe disease without simultaneously impacting XCI? Furthermore, if the SMCHD1 mutations did affect XCI, could XCI defects contribute to the presentation of arhinia and FSHD2? Here, we explore the genotype–phenotype relationships by analyzing transcriptomic data from human arhinia and FSHD2 patients, and document a surprising lack of XCI defects. However, we identify a link between SMCHD1 dysfunction and misregulation of select autosomal gene clusters that are candidate downstream targets for SMCHD1-related disorders.
Materials and Methods
RNA-sequencing analysis
RNA-sequencing (RNA-seq) data of female/male mouse embryonic stem (ES) cells (mESCs) (GSE54016) (Gendrel et al. 2014), female/male mouse liver cells (GSE48109) (Sugathan and Waxman 2013), male wild-type (WT)/Smchd1MommeD1/MommeD1 (Smchd1−/−) mouse neural stem/progenitor cells (NPCs) (GSE65747) (Chen et al. 2015), female WT/Smchd1−/− mouse NPCs (GSE99991) (Wang et al. 2018), and FSHD2 patients (GSE56787) (Yao et al. 2014) were downloaded from the Gene Expression Omnibus (GEO). Arhinia patient RNA-seq data have been published (Shaw et al. 2017). Reads were aligned to mm9 or hg19 using TopHat2 (Kim et al. 2013), with PCR duplicates removed from paired-end data sets. Strand information was resolved if the library was constructed using strand-specific protocols. Uniquely mapped exonic reads were counted using Homer (Heinz et al. 2010), and fragments per kilobase of transcript per million mapped reads (FPKMs)/reads per kilobase of transcript per million mapped reads (RPKMs) were calculated by dividing the RNA-seq exonic fragment/read count of each gene by the length of exons (kilobases) and total mapped exonic fragments/reads (million). Fold changes were calculated by dividing the FPKM/RPKMs of samples with SMCHD1 mutations with those of controls. To compare the fold changes of autosomal and X-linked genes, only genes with FPKM/RPKM > 0.1 were included. The lists of class I SMCHD1-senstive genes and genes subject to XCI in female mouse NPCs have been reported (Wang et al. 2018). To identify “facultative escapees,” we first compiled a list of all escapees reported from previous studies (Yang et al. 2010; Lopes et al. 2011; Splinter et al. 2011; Calabrese et al. 2012; Li et al. 2012; Pinter et al. 2012; Berletch et al. 2015; Marks et al. 2015) and defined facultative escapees as reported escapees that do not belong to “constitutive escapees” defined in female mouse NPCs (Wang et al. 2018). The human homologs of class I SMCHD1-senstive genes were identified based on a list of human–mouse homologs retrieved from the Mouse Genome Informatics database. We defined constitutive escapees (“E” or “Mostly E”), facultative escapees (“VE” or “Mostly VE”), and genes subject to XCI (“S” or “Mostly S”) in humans based on a previous study (Balaton et al. 2015), which comprehensively classified the XCI status of X-linked genes by compiling several existing studies. In the FSHD RNA-seq data sets, we excluded data of a control individual (C6) from our analyses as the library was sequenced at a much lower depth. Cumulative distribution plots were constructed using R and ggplot2. P-values were computed by Wilcoxon ranked sum test (unpaired, one-sided) using R. To visualize RNA-seq coverage, we generated fragment-per-million (fpm)-normalized bigWig files, which were displayed using Integrative Genomics Viewer (IGV) with scales indicated in each track.
To identify differentially expressed autosomal genes in Smchd1−/− female mouse NPCs, we analyzed RNA-seq data sets of two WT and two Smchd1−/− clones that carry two X chromosomes, together with those of one WT and one Smchd1−/− clone with XO genotypes, using the DESeq tool (Anders and Huber 2010). The inclusion of additional clones led to a reduced number of differentially expressed genes than that reported previously (Wang et al. 2018), as there was variation in gene expression between clones. Functional annotation clustering was performed using the Database for Annotation, Visualization and Integrative Discovery (DAVID) (Huang da et al. 2009), with stringency set as “medium.” Gene Ontology (GO) analysis was also performed using DAVID (Huang da et al. 2009). For functional annotation clustering and GO analysis, P-values corrected by the Benjamini–Hochberg method were presented.
To perform DESeq analyses for arhinia patients Y1 and B1, we compared each patient with 10 controls (A2, AE2, AE3, AE4, AH2, AH4, AH6, D2, D3, and D4). To perform DESeq analysis for FSHD2 patient F14, we compared F14 with eight controls (C1, C2, C3, C4, C5, C7, C8, and C9).
Chromatin immunoprecipitation-sequencing and in situ Hi-C data analysis
H3K4me3, H3K27me3, CCCTC-binding factor (CTCF), and RAD21 chromatin immunoprecipitation-sequencing (ChIP-seq) and in situ Hi-C data in mouse NPCs, derived by in vitro differentiation of WT and Smchd1−/− mESCs, were available from the GEO (GSE99991). EZH2 ChIP-seq data in mouse NPCs were uploaded to GSE126257. We displayed fpm-normalized ChIP-seq coverage “comp” (compiling allele-specific and nonallelic reads) tracks using IGV, with scales indicated in each track. Replicate 1 of ChIP-seq is shown in the figures, but replicate 2 demonstrated similar patterns at the Pcdh and Hox loci as replicate 1. Two biological replicates of in situ Hi-C data were pooled to increase depth. Combined Hi-C summary files were converted to .hic files, which were then displayed by Juicebox using the “balanced” mode for normalization (Durand et al. 2016), with resolutions and scales indicated in each panel. WT/Smchd1−/− maps were also computed by Juicebox. The hic files have been uploaded to the GEO (GSE126257).
Data availability
All next-generation sequencing data are available at the GEO with the accession numbers GSE54016, GSE48109, GSE65747, GSE99991, GSE56787, and GSE126257. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9633668.
Results
Dosage compensation is preserved in female arhinia and FSHD2 patients
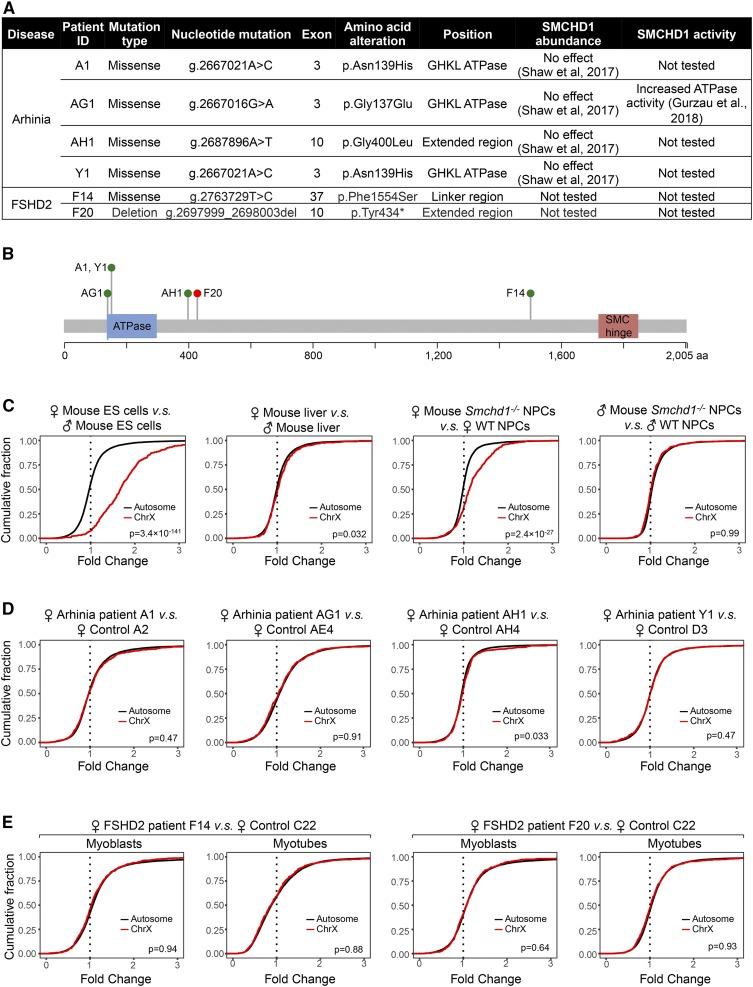
To determine if heterozygous mutations in SMCHD1 identified in patients with arhinia or FSHD2 are accompanied by defective XCI, we analyzed published RNA-seq data sets in lymphoblastoid cell lines (LCLs) from four female arhinia patients, myogenic cells from two female FSHD2 patients (Figure 1, A and B), and corresponding cell types from female controls. Arhinia patients carry heterozygous missense SMCHD1 mutations that are clustered at the N-terminal ATPase domain of the protein (Gordon et al. 2017; Shaw et al. 2017), whereas FSHD2 patients harbor missense, nonsense, and deletion mutations spanning the entire SMCHD1 locus (Figure 1, A and B) (Lemmers et al. 2012; Jansz et al. 2017; Shaw et al. 2017). The interrogation of individual X-linked genes with this small sample size and limited number of replicates posed a challenge because defective XCI typically leads to upregulation of any given X-linked gene by only up to twofold (Gendrel et al. 2012; Mould et al. 2013; Yang et al. 2016; Wang et al. 2018). To overcome this technical limitation, we compared the cumulative distribution of the fold change of X-linked genes relative to all autosomal genes in female patients and controls (Yang et al. 2016; Carrette et al. 2018), reasoning that even a small degree of upregulation of many X-linked genes would cause a significant deviation in the X-linked gene fold change distribution compared with the distribution for autosomal genes.
Figure 1.
Dosage compensation is preserved in female arhinia and FSHD2 patients harboring SMCHD1 mutations. (A) A table listing the cohort of female arhinia and FSHD2 patients, and their associated SMCHD1 mutations included in this study. Arhinia patients harboring SMCHD1 mutations often exhibit normal levels of SMCHD1 protein (Shaw et al. 2017). In contrast, it has been reported that SMCHD1 mutations in FSHD2 patients are usually associated with reduced SMCHD1 protein levels (Lemmers et al. 2012), although the two FSHD2 patients listed here have not been tested for SMCHD1 levels. (B) The domain structure of human SMCHD1 protein with the locations of mutations found in patients listed in (A) overlaid. Green dots, missense mutations. Red dot, nonsense or frameshift mutations. This figure was modified from Shaw et al. (2017). (C) CDPs for fold changes of X-linked and autosomal genes in mouse cells. Note that X–A imbalance was not seen in male Smchd1−/− mouse NPCs, indicating that X–A imbalance in Smchd1−/− females originated from the inactive X chromosome. P-values given by Wilcoxon ranked sum test (unpaired, one-sided). (D) CDPs for fold changes of X-linked and autosomal genes in female arhinia patients vs. female controls. (E) CDPs for fold changes of X-linked and autosomal genes in female FSHD2 patients vs. female controls. CDP, cumulative distribution plot; Chr, chromosome; ES, embryonic stem; FSHD2, facioscapulohumeral muscular dystrophy type 2; ID, identifier; NPC, neural progenitor cell; SMC, structural maintenance of chromosomes; SMCHD1, SMC flexible hinge domain-containing 1; GHKL, Gyras, Hsp90, Histidine Kinase, MutL; WT, wild-type.
To validate the sensitivity and specificity of our analytical pipeline for detecting dosage imbalances, we ran the analysis on mouse cells for which a dosage imbalance has been established. First, we examined publicly available RNA-seq data from XX and XY mESCs (Gendrel et al. 2014). Undifferentiated female mESCs have two active X chromosomes (Xa), whereas male mESCs have one Xa. Indeed, we observed a rightward shift of X-linked fold change distribution relative to autosomes between female and male mESCs (Figure 1C, left), indicating disproportionate upregulation of X-linked genes relative to autosomes in females. This “X–A imbalance” was absent, as expected, between WT male and female mouse hepatocytes (Sugathan and Waxman 2013), as these fully differentiated cells had undergone XCI. (Figure 1C, left center). We also observed X–A imbalance in Smchd1−/− NPCs derived from female mESCs (Figure 1C, right center) (Wang et al. 2018) but not male mice (Figure 1C, right) (Chen et al. 2016). These analyses demonstrate the robustness of our analytical pipeline.
We next examined RNA-seq data from female arhinia and FSHD2 patients. We analyzed each patient separately, reasoning that each female patient harbors a distinct mutation that may affect SMCHD1 differently (Figure 1A). Indeed, recent work has shown varying ATPase activity levels in distinct SMCHD1 mutants (Gurzau et al. 2018). Examining each patient separately, we found that none of the four female arhinia patients with heterozygous missense mutations in SMCHD1 demonstrated X–A imbalance (Figure 1D). Interestingly, X upregulation was also not detected in either female FSHD2 patient (Figure 1E), despite one of them (F20) harboring a frameshifting deletion in SMCHD1. Thus, these specific SMCHD1 mutations do not cause dosage compensation defects, at least when examined on a global chromosomal scale in heterozygous human patients.
Class I genes resist arhinia- and FSHD2-associated SMCHD1 mutations
Previous studies showed that in mice, SMCHD1 is required to silence a specific subset of genes on the Xi, referred to as class I SMCHD1-sensitive genes (class I genes, n = 126) (Wang et al. 2018). In the absence of SMCHD1, class I genes exhibited local Xist RNA-spreading defects and erosion of heterochromatic silencing, while other genes on the Xi were not affected. Overall, Smchd1 ablation leads to defective silencing in ≤ 43% of genes normally subject to XCI (Gendrel et al. 2013; Mould et al. 2013; Wang et al. 2018). Indeed, the X–A imbalance between Smchd1−/− and WT mouse NPCs was weaker than that between female and male mESCs (compare Figure 1C, left and right center), indicating a partial XCI defect. We reasoned that an X–A imbalance in these patients may not be evident if only a small fraction of X-linked genes were dysregulated. To address this possibility, we tested if a weaker X–A imbalance could be detected in patients by focusing on the subset of genes that are known to be dependent on SMCHD1 for silencing.
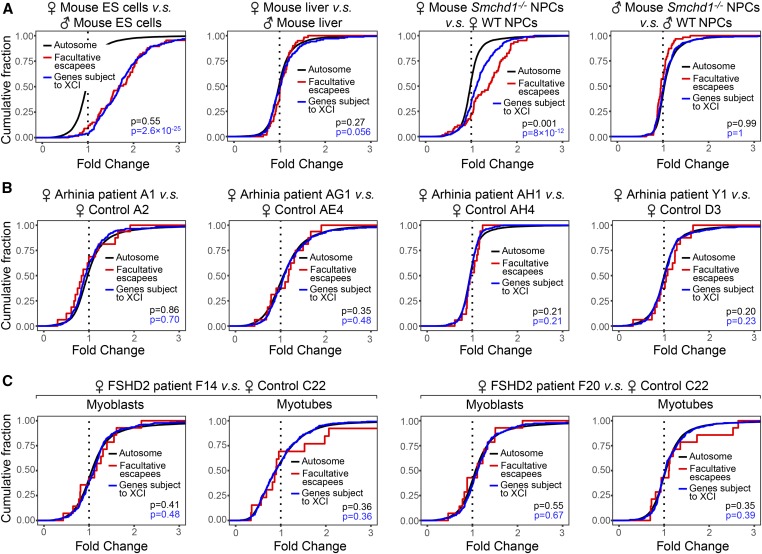
To validate the approach, we first compared the expression of “escapees,” a subset of X-linked genes resistant to XCI, between female and male controls (Supplemental Material, Figure S1). We focused on constitutive escapees, which resist XCI in all cells (Berletch et al. 2011; Balaton and Brown 2016). When examining fold change distribution upon aggregating all X-linked genes, the increased dosage of constitutive escapees in females led to a minimal X–A imbalance between females and males (Figure S1A), which was not seen between same-sex individuals (Figure S1, B and C). Notably, when analyzing constitutive escapees (n = 49) separately from other X-linked genes, we detected upregulation of these escapees in females relative to males (Figure S1D), but not between individuals of the same sex (Figure S1, E and F). Thus, X–A imbalance can be sensitively detected by examining select X-linked genes with increased dosage.
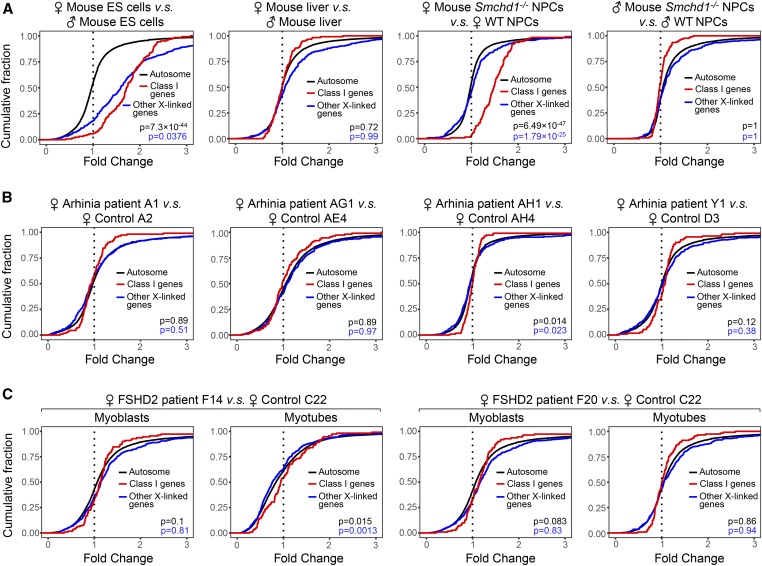
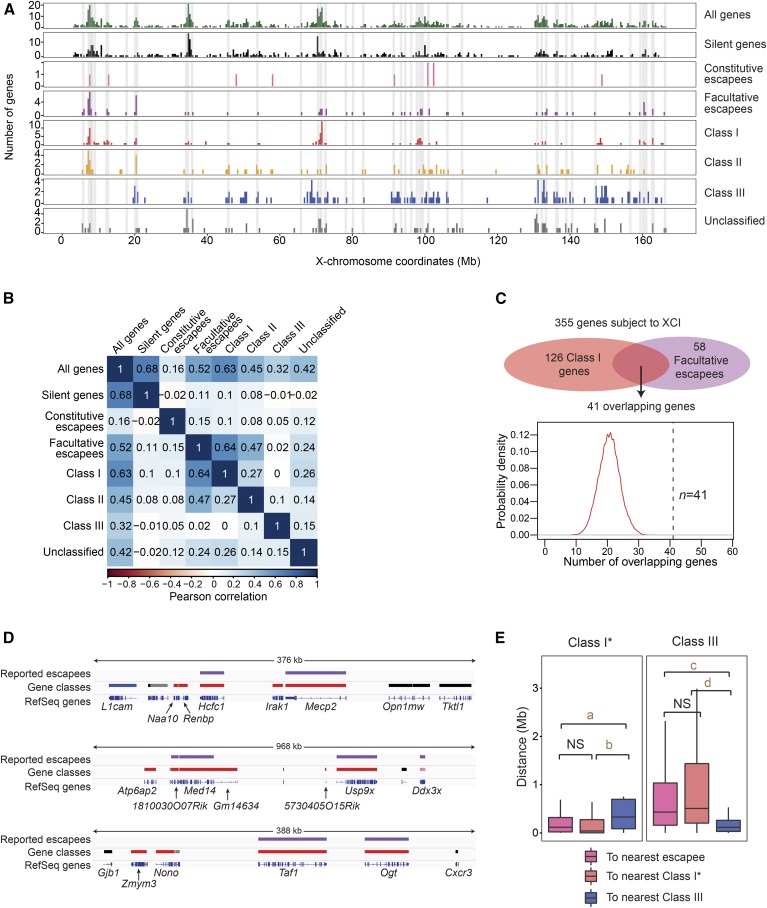
We next focused analyses on human X-linked genes homologous to mouse class I genes (n = 116). Significantly, female arhinia patients (n = 4) and FSHD2 patients (n = 2) did not demonstrate upregulation of class I genes compared to female controls (Figure 2, B and C). In mice, the unique dependence of class I genes on SMCHD1 for silencing is intriguing. Because SMCHD1 binds broadly along the entire Xi in mice (Wang et al. 2018), the specific effect on class I genes cannot be conferred by region-specific SMCHD1 binding. We asked if geographic characters on the Xi may affect regional SMCHD1 sensitivity. It has been reported that class I genes form clusters (Gendrel et al. 2013; Wang et al. 2018). Interestingly, class I gene clusters overlapped with regions known to be rich in facultative escapees (r = 0.64) (Figure 3, A and B), a distinct subset of escapees that resist XCI only in select cells or tissues (Berletch et al. 2011; Balaton and Brown 2016) (Table S1). Furthermore, facultative escapees were selectively enriched in class I genes (P < 0.0001) (Figure 3C), and nonescaping class I genes tended to be in the vicinity of escapees (Figure 3, D and E), suggesting that regions harboring facultative escapees may be more sensitive to Smcdh1 dysfunction. Indeed, facultative escapees were upregulated in Smchd1−/− female mouse NPCs (Figure 4A). Our findings in mice are in line with a study that links genetic variants of SMCHD1 to facultative escapees in humans (Luijk et al. 2018). In spite of this, we observed no upregulation of facultative escapees (n = 44) in female arhinia (n = 4) or FSHD2 (n = 2) patients (Figure 4, B and C). Collectively, these data indicate that arhinia- and FSHD2-assocated SMCHD1 mutations do not affect dosage compensation of X-linked genes when they are in the heterozygous state. Thus, dosage compensation is less sensitive to SMCHD1 dysfunction than are craniofacial development and muscular function.
Figure 2.
Human homologs of class I SMCHD1-sensitive Xi genes resist upregulation in heterozygous female patients. (A) CDPs comparing class I SMCHD1-sensitive genes (class I genes), other X-linked genes, and autosomal genes in mouse cells. P-values: class I genes vs. autosomal genes (black); class I genes vs. other X-linked genes (blue). P-values given by Wilcoxon ranked sum test (unpaired, one-sided). (B) CDPs comparing human homologs of class I SMCHD1-sensitive genes (class I genes), other X-linked genes, and autosomal genes in female arhinia patients vs. female controls. (C) CDPs comparing human homologs of class I SMCHD1-sensitive genes (class I genes), other X-linked genes, and autosomal genes in female FSHD2 patients vs. female controls. CDP, cumulative distribution plot; ES, embryonic stem; FSHD2, facioscapulohumeral muscular dystrophy type 2; NPC, neural progenitor cell; SMCHD1, structural maintenance of chromosomes flexible hinge domain-containing 1; WT, wild-type; Xi, inactive X chromosome.
Figure 3.
Inactivation of facultative escapees is sensitive to Smchd1 ablation in mice. (A) Location of facultative escapees relative to various X-linked gene classes. Gray-shaded areas are regions with facultative escapees. (B) Pearson correlation analysis of the occurrence of facultative escapees relative to various X-linked gene classes on the X. (C) Testing the random likelihood of having 41 class I genes as facultative escapees. Random samplings of 126 genes from 355 genes subject to XCI in which 58 are facultative escapees were simulated 10,000 times. A probability density plot was generated for the number of facultative escapees sampled. (D) Representative examples of class I genes that either overlap with or are adjacent to reported escapees. Top track: all reported (constitutive and facultative) escapees (purple bars). Middle track: silent genes (black bars), constitutive escapees (pink bars), class I genes (red bars), unclassified genes (gray bars), and class III genes (blue bars). Bottom track: RefSeq genes. (E) Nearest neighbor analysis: box plots showing distribution of distances of class I and class III genes from each other, and to escapees. e.g., the left panel shows the distance distribution from a class I gene to the nearest escapee (pink), class I (red), or class III (blue) gene. For this analysis, the 41 facultative escapees are removed from the class I list (class I*). P-values were determined by Wilcoxon ranked sum test. NS, not significant (P > 0.05). a, P = 3.303 × 10−4. b, P = 3.221 × 10−6. c, P = 6.14 × 10−12. d, P = 8.556 × 10−15. XCI, X-chromosome inactivation.
Figure 4.
Behavior of facultative escapees in female arhinia and FSHD2 patients. (A) CDPs comparing facultative escapees, genes subject to XCI, and autosomal genes in mouse cells. P-values: facultative escapees vs. autosomal genes (black); facultative escapees vs. genes subject to XCI (blue). P-values given by Wilcoxon ranked sum test (unpaired, one-sided). (B) CDPs comparing facultative escapees, genes subject to XCI, and autosomal genes in female arhinia patients vs. female controls. (C) CDPs comparing facultative escapees, genes subject to XCI, and autosomal genes in female FSHD2 patients vs. female controls. CDP, cumulative distribution plot; ES, embryonic stem; FSHD2, facioscapulohumeral muscular dystrophy type 2; NPC, neural progenitor cell; WT, wild-type; XCI, X-chromosome inactivation.
SMCHD1 controls the epigenetic state and 3D organization of the mouse Pcdh cluster
It is believed that SMCHD1 mutations, in conjunction with a shortened permissive D4Z4 array, cause FSHD2 via derepression of the leading FSHD candidate gene, Double Homeobox 4 (DUX4) (Lemmers et al. 2012), but other unknown genetic (or) environmental factors may also be contributory (Jones et al. 2012). The molecular basis of arhinia remains unclear, but our previous transcriptome studies suggested DOK7, TGIF1, KDM6A, and ICK to be dysregulated in arhinia patients (Shaw et al. 2017). To gain insight into the pathogenesis of arhinia and FSHD2, we investigated the effect of SMCHD1 dysfunction on autosomal gene expression. We began with an examination of autosomal genes dysregulated in Smchd1−/− mouse cells, reasoning that the stronger effect of homozygous mutations may allow a more targeted approach in analyzing the patient data, where heterozygous mutations may only yield subtle changes in gene expression.
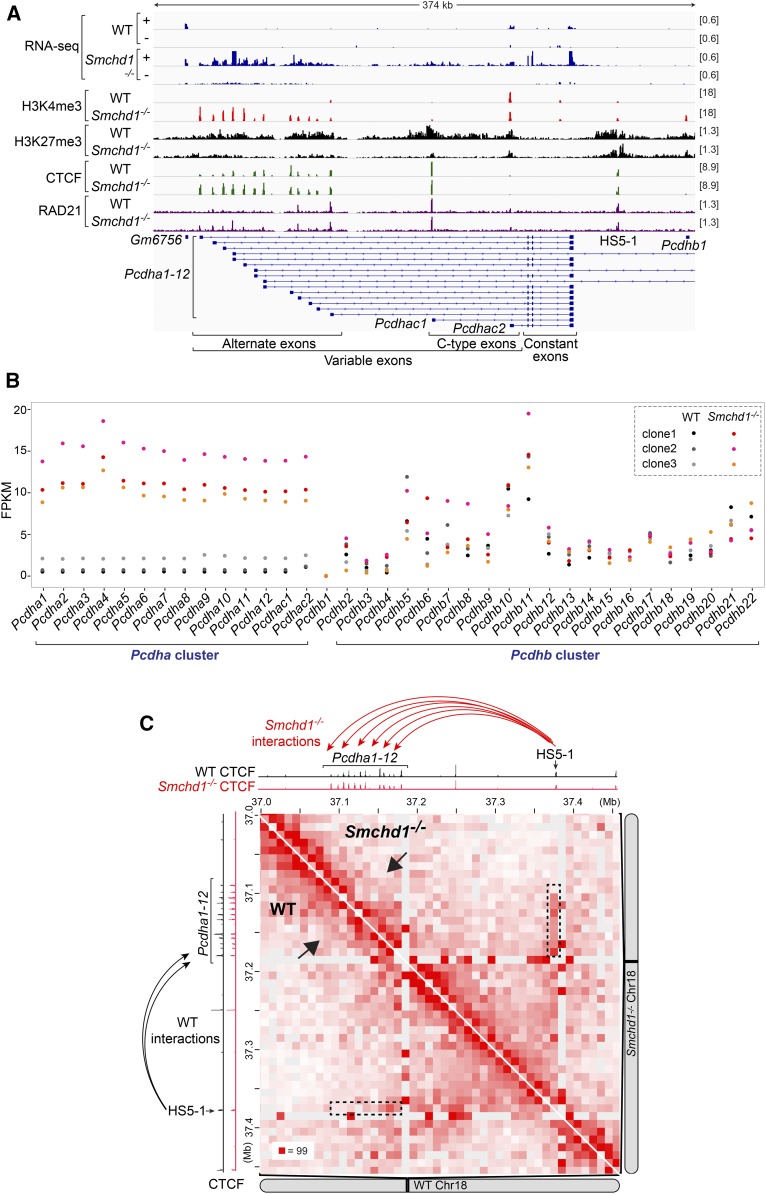
It is known that Smchd1−/− mice exhibit upregulation and CpG hypomethylation of the clustered protocadherin (Pcdh) genes (Gendrel et al. 2012; Mould et al. 2013; Chen et al. 2015; Wang et al. 2018). These genes encode a subgroup of cadherin proteins critical for neuronal diversity and neural circuit formation (Zipursky and Sanes 2010; Chen and Maniatis 2013; Yagi 2013; Mountoufaris et al. 2017). The Pcdh genes are arranged into three closely linked clusters—α, β, and γ—with dysregulation of the Pcdha cluster in the Smchd1−/− background being most consistently observed. Each Pcdha mRNA contains one of the 14 tandemly arranged “variable exons,” with each carrying a promoter, and three “constant exons” shared by all Pcdha isoforms (Figure 5A). Through stochastic promoter choice, Pcdha isoforms carrying the 12 “alternate” exons exhibit combinatorial and monoallelic expression in neuronal lineages, whereas isoforms carrying the two “C-type” exons are expressed biallelically and constitutively (Wu and Maniatis 1999; Wu et al. 2001; Tasic et al. 2002; Wang et al. 2002; Esumi et al. 2005). To probe deeper into how the Pcdh genes are aberrantly regulated in the absence of SMCHD1, we reanalyzed published RNA-seq data in WT (n = 3) and Smchd1−/− (n = 3) female mouse NPC clones, which were derived from in vitro differentiation of WT and Smchd1−/− mESCs (Wang et al. 2018). In all three Smchd1−/− clones, we observed upregulation of all 14 Pcdha isoforms (Figure 5B, left), whereas the expression of Pcdhb (Figure 5B, right) and Pcdhg clusters (Figure S2A) was similar between WT and Smchd1−/− clones. Strand-specific RNA-seq coverage profiles (Figure 5A, “+” strand) confirmed Pcdha upregulation. Interestingly, we also observed increased antisense transcription at regions harboring alternate exons in Smchd1−/− cells (Figure 5A and Figure S2B, “−” strand).
Figure 5.
Smchd1 ablation alters the expression, chromatin states, and three-dimensional organization of the Pcdha gene cluster in mice. (A) RNA-seq, H3K4me3, H3K27me3, CTCF, and RAD21 ChIP-seq (GSE99991) tracks at the Pcdha cluster in WT (clone1) and Smchd1−/− (clone1) female mouse NPCs, with scales indicated in each track. +, the plus strand. −, the minus strand. (B) Dot plots showing the FPKMs of the 14 Pcdha and 22 Pcdhb genes in WT (n = 3) and Smcdh1−/− (n = 3) mouse NPC clones. (C) Hi-C contact maps at 10-kb resolution at the Pcdhα cluster in WT (clone1, bottom) and Smchd1−/− (clone1, top) female mouse NPCs (GSE99991). Also shown are CTCF ChIP-seq tracks in WT (black) and Smchd1−/− (red) female mouse NPCs. ChIP-seq, chromatin immunoprecipitation-sequencing; Chr, chromosome; CTCF, CCCTC-binding factor; FPKM, fragments per kilobase of transcript per million mapped reads; NPC, neural progenitor cell; RNA-seq, RNA-sequencing; WT, wild-type.
We then investigated changes at the Pcdh locus at the level of chromatin using published epigenomic profiling data (Wang et al. 2018). In the WT clone, only one alternate (Pcdha12) and two C-type exons (Pcdhac1 and Pcdhac2) were marked by the active H3K4me3 histone mark (Figure 5A) [note that in WT clones shown in Figure 5B, Pcdha12, Pcdhac1, and Pcdhac3 did not show greater expression (FPKM) than other isoforms, as reads mapped to the constant exons were counted for all Pcdhas]. In Smchd1−/− cells, all variable exons became H3K4me3-enriched, coinciding with depletion of the repressive H3K27me3 histone mark and aberrant expression of all Pcdha isoforms (Figure 5, A and B), consistent with a previous report (Chen et al. 2015).
Transcription of Pcdha genes is regulated by hypersensitivity site 5-1 (HS5-1), an enhancer element located downstream of the last constant exon (Figure 5A) (Ribich et al. 2006; Kehayova et al. 2011). HS5-1 interacts with Pcdha promoters via long-range DNA looping mediated by two architectural factors, CTCF and cohesin (Guo et al. 2012; Monahan et al. 2012). We found that in Smchd1−/− mouse NPCs, activation of Pcdha1-11 was associated with increased binding of not only CTCF [as previously reported (Chen et al. 2015)] but also RAD21, a cohesin subunit, to their respective promoters (Figure 5A and Figure S2B). We then examined in situ Hi-C data sets in WT and Smchd1−/− mouse cells (Wang et al. 2018) to determine if enhanced CTCF/cohesin binding is accompanied by altered 3D topological organization of this locus. Strikingly, chromatin interactions between Pcdha1-12 promoters and the HS5-1 enhancer became more frequent in Smchd1−/− than those in WT cells (Figure 5C, dashed rectangles), although the limited resolution of the Hi-C data sets did not allow us to pinpoint contacts between HS5-1 and each individual Pcdha1-12 promoter. Moreover, increased CTCF/cohesin binding at the Pcdha5 promoter in Smchd1−/− cells coincided with a strengthened border that appeared to separate Pcdha1-12 into two small interacting domains (Figure 5C, arrows). Thus, our analyses revealed that SMCHD1 regulates not only the expression, but also the chromatin states and 3D organization, of the Pcdha gene cluster in mice.
Aberrant expression of PCDHA genes in arhinia and FSHD2 patients
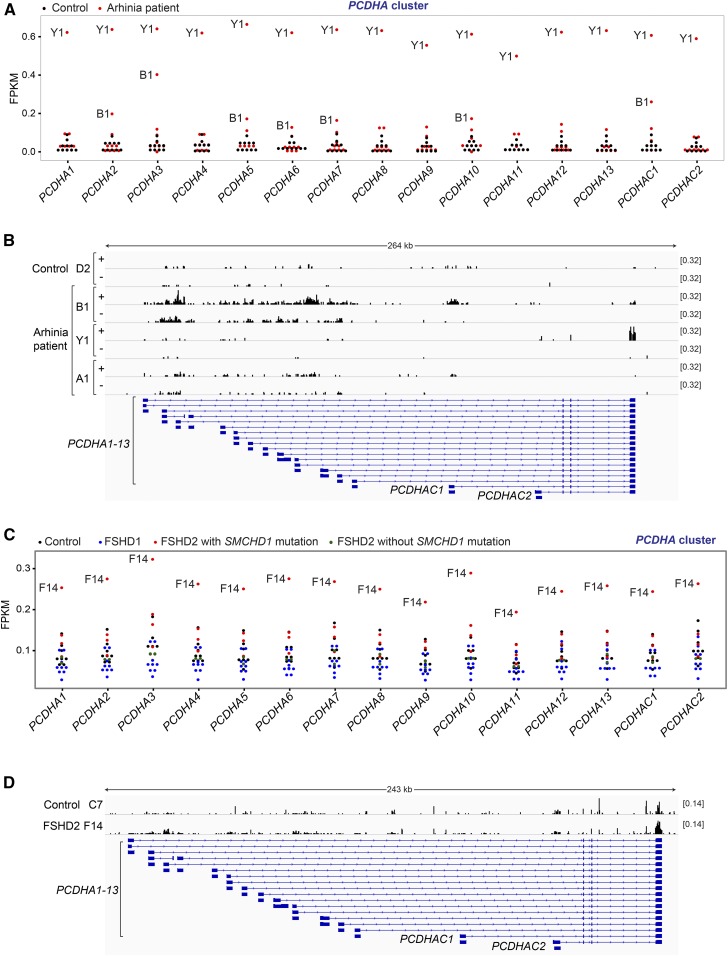
Given effects on the mouse Pcdha locus, we asked if human PCDH clusters are affected by the arhinia and FSHD2-associated SMCHD1 mutations. While the PCDH genes are predominantly expressed in the nervous system, clustered Pcdh/PCDH upregulation has been detected in nonneuronal lineages, such as in Smchd1−/− whole-mouse embryos (Gendrel et al. 2013; Mould et al. 2013), Smchd1−/− mouse embryonic fibroblasts (Leong et al. 2013), and FSHD2 human myogenic cells (Yao et al. 2014; Mason et al. 2017). With this in mind, we examined PCDH expression in RNA-seq data of LCLs from four females (Figure 1A) and six males (Figure S3A) with arhinia, with 10 unaffected individuals serving as controls. Interestingly, we observed upregulation of PCDHA genes in two arhinia patients (B1 and Y1) (Figure 6A and Table S2), but not in any of the controls. On the other hand, the PCDHB cluster was not expressed in LCLs even in normal cells (FPKM < 0.1), and the PCDHG cluster was not differentially expressed in patient vs. normal cells (Figure S3B).
Figure 6.
Dysregulation of the PCDHA gene cluster in arhinia and FSHD2 patients. (A) Dot plots showing the FPKMs of the 15 PCDHA genes in LCLs of 10 arhinia patients (red) and 10 controls (black). See Table S2 for the results of DESeq analyses, which similarly showed upregulation of PCDHA genes in patient Y1 and B1. (B) Strand-resolved RNA-seq coverage tracks at the PCDHA gene cluster of three arhinia patients (B1,Y1, and A1) and one control (D2). +, the plus strand. −, the minus strand. (C) Dot plots showing the FPKMs of the 14 PCDHA genes in muscle biopsies of nine FSHD1 patients (blue), four FSHD2 patients carrying SMCHD1 mutations (red), two FSHD2 patients without SMCHD1 mutations (green), and eight controls (black). See Table S2 for the results of DESeq analyses, which similarly showed upregulation of PCDHA genes in patient F14. (D) RNA-seq coverage tracks at the PCDHA gene cluster of one FSHD2 patients (F14) and one control (C7). +, the plus strand. −, the minus strand. FPKM, fragments per kilobase of transcript per million mapped reads; FSHD2, facioscapulohumeral muscular dystrophy type 2; LCL; lymphoblastoid cell line; RNA-seq, RNA-sequencing; SMCHD1, structural maintenance of chromosomes flexible hinge domain-containing 1.
Therefore, we focused on PCDHA and inspected strand-specific RNA-seq coverage tracks. We observed distinct patterns of PCDHA derepression in these two arhinia patients. In patient B1, most RNA-seq reads of the PCDHA genes originated from variable exons, but many reads were mapped to the intronic regions between alternate exons (Figure 6B, “+” strand) [note that in Figure 6A, B1 did not appear to be upregulated as strongly as what was shown in Figure 6B because only exonic reads were counted in the FPKM analysis). Furthermore, PCDHA derepression in B1 was associated with enhanced antisense transcription (Figure 6B, “−” strand), reminiscent of the pattern seen in Smchd1−/− mouse NPCs (Figure 5A). This distinct profile was detected in B1, but not in any other arhinia patients (n = 9) or controls (n = 10) (Figure 6B). Notably, the intronic and antisense PCDHA reads did not stem from artifacts of library construction and/or sequencing, as the RNA-seq of patient B1 at other loci exhibited characteristic exonic enrichment in the sense orientation (Figure S3C), and were sequenced to a depth similar to other patients/controls.
In patient Y1, PCDHA RNA-seq reads originated predominantly from the last constant exon (Figure 6B). All PCDHA isoforms were therefore counted as upregulated in the FPKM analysis (Figure 6A). We noted that patient A1, who carried the same missense mutation (p.Asn139His) as Y1, did not exhibit PCDHA upregulation (Figure 6B). These data imply that this SMCHD1 mutation is not sufficient to cause misexpression of the PCDHA cluster; there are likely modifier genes that aid suppression of PCDHA in the presence of the SMCHD1 missense mutation.
We also examined clustered PCDH expression in four FSHD2 patients carrying SMCHD1 mutations using RNA-seq data of their muscle biopsies (Yao et al. 2014). Eight healthy individuals (controls), nine FSHD1 patients, and two FSHD2 patients without SMCHD1 mutations were also analyzed (Yao et al. 2014). Expression of all three PCDH clusters was detected in muscle biopsies. In one FSHD2 patient (F14), we found upregulation of all PCDHA and three PCDHB isoforms (Figure 6C, Figure S4A, and Table S2), but not the PCDHG genes (Figure S4B). RNA-seq coverage profiles also revealed increased read density at the PCDHA loci in F14 (Figure 6D); however, we could not evaluate antisense transcription as the libraries were constructed using a nonstrand-specific protocol. PCDHA upregulation was not detected in other FSHD1 and FSHD2 patients, suggesting an effect specific to this SMCHD1 mutation and/or other factors unique to F14. In vitro-cultured myotubes from F14 also exhibited PCDHA upregulation (Figure S5), although there was considerable variation in the expression levels of PCDHA in control myotubes. Taken together, our observation of PCDHA upregulation in several arhinia and FSHD2 patients suggests that SMCHD1 may regulate this gene cluster in humans, as in mice. Genetic background and modifier loci can clearly also influence PCDHA expression when SMCHD1 activity is aberrant.
Smchd1 ablation causes aberrant regulation of Hox genes in mice
To identify additional classes of autosomal genes controlled by SMCHD1, we performed functional annotation clustering on autosomal genes differentially expressed in Smchd1−/− mouse NPCs (Tables S3 and S4) (Wang et al. 2018). Strikingly, among the upregulated autosomal genes (n = 251), we found homeobox genes, a group of transcription factors that control early embryonic development, to be one of the most highly enriched functional clusters (enrichment score = 5.4; P = 2.6 × 10−11), second only to the protocadherin genes (enrichment score = 6.39; P = 3.9 × 10−7) (Table S5). In contrast, no functional clusters in downregulated genes (n = 82) achieved statistical significance (Table S6).
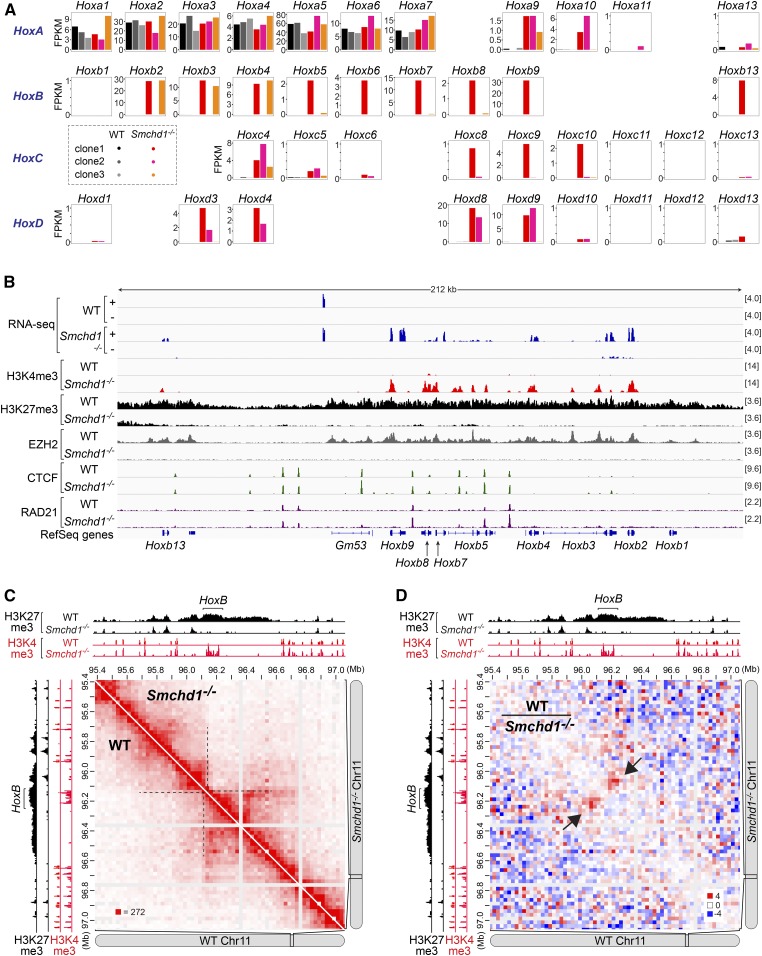
Around 67% (10/15) of the homeobox genes derepressed in Smchd1−/− mouse NPCs are members of the Hox gene family, which controls the body plans in metazoans (Lewis 1978; Izpisúa-Belmonte et al. 1991; Duboule 2007). Hox genes, like the Pcdh genes, are clustered. There are four Hox clusters in vertebrates: HoxA, HoxB, HoxC, and HoxD (Duboule and Dolle 1989; Graham et al. 1989). The ordering of Hox genes within each cluster matches their unique spatiotemporal expression pattern during embryonic development, with genes at the 3′ end of the cluster being activated earlier and along the length of the animal (“anterior Hox genes”), and genes at the 5′ end being expressed later and in posterior structures (“posterior Hox genes”) (Lewis 1978; Izpisúa-Belmonte et al. 1991; Duboule 2007). In three WT mouse NPC clones, we observed the same pattern of Hox expression, with only the anterior portion of HoxA (Hoxa1-7) being activated and the others remaining repressed (Figure 7A, black and gray bars), indicative of the rostral identity of these cells. While Hoxa1-7 remained active in the absence of SMCHD1, all three Smchd1−/− clones exhibited aberrant Hox derepression (Figure 7A). Interestingly, in contrast to the uniform upregulation of the Pcdha cluster (Figure 5B), derepression of Hox genes was heterogeneous. Aberrant Hox activation appeared in all four paralogous clusters, but the number of activated genes varied not only between Hox clusters but also between different NPC clones (Figure 7A). Furthermore, we detected derepression not only of members of the Hoxa1-7 paralogy groups, but also posterior Hox genes such as Hox9-10, indicative of caudal/posterior transformation.
Figure 7.
Smchd1 ablation alters the expression, chromatin states, and three-dimensional organization of the HoxB gene cluster in mice. (A) Bar plots showing the FPKMs of all four clusters of Hox genes in WT (n = 3) and Smcdh1−/− (n = 3) mouse NPC clones. (B) RNA-seq, H3K4me3, H3K27me3, EZH2, CTCF, and RAD21 ChIP-seq (GSE99991) tracks at the HoxB cluster in WT (clone1) and Smchd1−/− (clone1) female mouse NPCs, with scales indicated in each track. +, the plus strand. −, the minus strand. (C) Hi-C contact maps at 25-kb resolution at the HoxB cluster in WT (clone1, bottom) and Smchd1−/− (clone1, top) female mouse NPCs (GSE99991). Also shown are H3K4me3 and H3K27me3 ChIP-seq tracks in WT and Smchd1−/− female mouse NPCs. (D) Differential Hi-C contact maps generated by dividing the WT with the Smchd1−/− contact map at 25-kb resolution at the HoxB cluster (GSE99991). ChIP-seq, chromatin immunoprecipitation-sequencing; CTCF, CCCTC-binding factor; FPKM, fragments per kilobase of transcript per million mapped reads; NPC, neural progenitor cell; RNA-seq, RNA-sequencing; WT, wild-type.
Hox genes are regulated by Polycomb group proteins, a set of repressive histone-modifying complexes that control developmental genes (Schwartz and Pirrotta 2007; Soshnikova and Duboule 2009; Surface et al. 2010). Indeed, the derepressed Hox loci were accompanied by decreased occupancy of EZH2, the catalytic subunit of Polycomb Repressive Complex 2 (PRC2), and its catalytic product, the H3K27me3 histone mark, in all four Hox clusters (Figure 7B, HoxB; Figure S6, HoxA and HoxD; and Figure S7A, HoxC). These derepressed genes were instead enriched by H3K4me3. Ablating Smchd1 is known to influence the binding of CTCF and cohesin (Chen et al. 2015; Wang et al. 2018; Gdula et al. 2019), as exemplified by the clustered Pcdha locus (Figure 5A). Indeed, altered binding intensity of CTCF and RAD21 at the Hox loci was occasionally observed at some sites (Figure 7B, and Figures S6 and S7A), but globally they did not bind more strongly compared to what was observed at the Pcdha1-11 region (Figure 5A). This difference might reflect different regulatory mechanisms at play at these loci and also varying degrees of derepression in Smchd1−/− cells.
The Hox clusters reside at the borders between topologically associated domains (TADs), megabase-sized chromatin structures that insulate interactions between loci from different domains (Dixon et al. 2012; Nora et al. 2012) or submegabase-scale TAD-like structures (subTADs). This unique positioning at TAD/subTAD boundaries contributes to sequential Hox activation (Dixon et al. 2012; Andrey et al. 2013; Berlivet et al. 2013; Darbellay and Duboule 2016). For instance, the HoxB cluster lies between two subTADs on mouse chromosome 11 (Figure 7C, dashed lines), with the boundary at the region between Hoxb9 and Hoxb13. Intriguingly, we examined in situ Hi-C data sets in Smchd1−/− NPCs (Wang et al. 2018) and found altered 3D organization at this subTAD boundary. The inter-subTAD interactions became decreased when Smchd1 was ablated (Figure 7C). To better visualize altered chromatin structures, we generated a differential Hi-C interaction map by dividing the WT by the Smchd1−/− map, which revealed more frequent inter-subTAD interactions in WT cells than in Smchd1−/− cells (Figure 7D, arrows). Altered inter-subTAD interactions were also observed for the boundary housing the HoxC cluster (between Hoxc9 and Hoxc13) (Figure S7, B and C, arrows), but not for the boundary downstream to HoxC (Figure S7, B and C, arrow heads), indicating an effect specific to Hox-associated boundaries. As Polycomb complexes are known to promote formation of self-interacting domains of compacted chromatin (“Polycomb domains”) (Eskeland et al. 2010; Bantignies et al. 2011; Schoenfelder et al. 2015; Kundu et al. 2017), the attenuated inter-subTAD interactions might be attributable to weakening of Polycomb domains that normally span TAD/subTAD boundaries and/or from altered CTCF/cohesin binding near these borders.
Taken together, these data demonstrate that SMCHD1 plays a critical role in the proper regulation of the Hox clusters in mice. Overall, the negative effects of Smchd1 ablation on Polycomb recruitment and the positive effects on H3K4me3, CTCF, and cohesin enrichment mirror what have been observed on the Xi (Jansz et al. 2018a; Sakakibara et al. 2018; Wang et al. 2018; Gdula et al. 2019). The accompanying changes in Hox domain topology are also consistent with a role for SMCHD1 in organizing 3D chromosomal structures. Thus, SMCHD1 plays a vital role on the X chromosome and select loci on autosomes in the mouse.
HOX gene regulation is preserved in LCLs of arhinia patients and muscle biopsies in FSHD2 patients
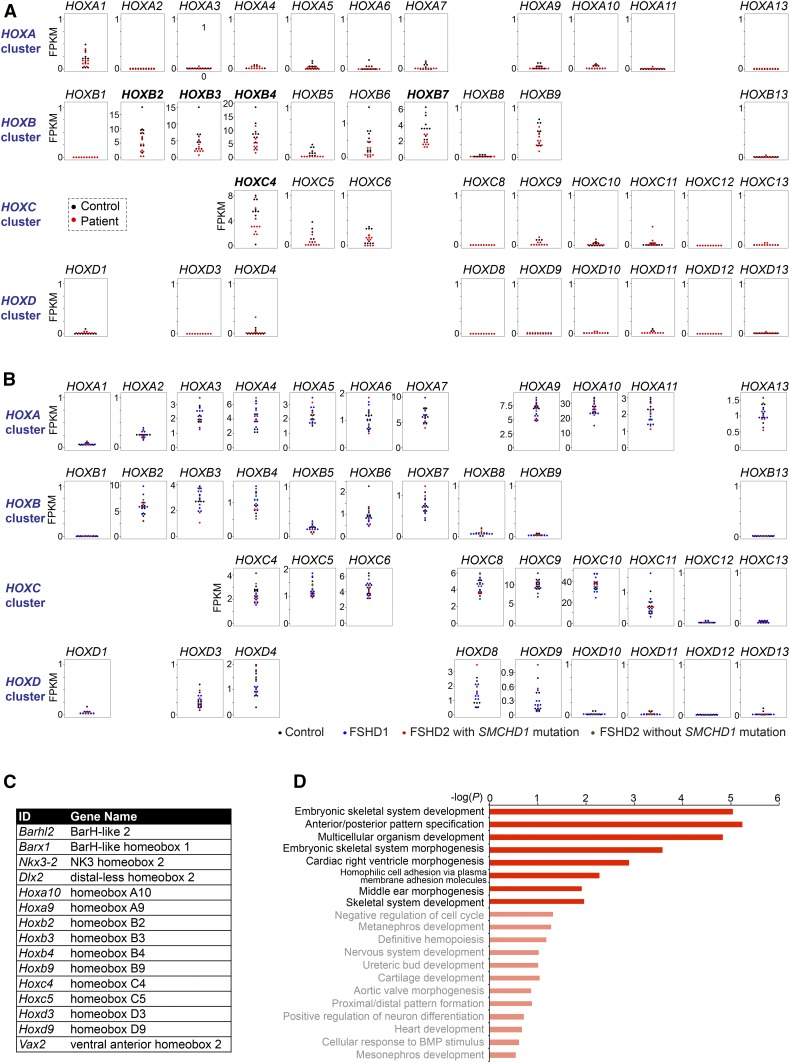
A role for SMCHD1 in controlling the mouse Hox genes is intriguing. Congenital arhinia often meets the diagnostic criteria of Bosma arhinia microphthalmia syndrome (Bosma et al. 1981) and is accompanied by other craniofacial abnormalities, such as cleft lip/palate, low-set ears, anophthalmia or microophthalmia, uveal colomoma, and hypogonadotropic hypogonadism. These phenotypes likely represent a developmental defect in cranial placodes and/or neural crest cells (NCCs) (Gordon et al. 2017; Shaw et al. 2017). In light of the critical role of Hox genes in regulating the specification and migration of cranial NCCs during development [reviewed in Parker et al. (2018)], our finding suggests that SMCHD1 mutations might cause arhinia and its associated craniofacial defects via dysregulation of the Hox genes. For instance, cranial NCCs derived from the anterior-most region of the neural tube, which normally lack Hox expression (Krumlauf 1993), may be susceptible to ectopic Hox expression when SMCHD1 is dysfunctional. The posterior transformation observed in Smchd1−/− mouse NPCs also supports speculation that rostral structures, such as derivatives of cranial NCCs, might be susceptible to phenotypic alteration associated with SMCHD1 dysfunction. With this in mind, we investigated HOX gene expression in the RNA-seq data of arhinia patients, noting that the data were from LCLs and not NCCs. We detected expression of HOXB2-4, HOXB7, and HOXC4 in the LCLs of control individuals (n = 10) (Figure 8A, black dots). However, this expression pattern was unchanged in LCLs of arhinia patients (n = 10) (Figure 8A, red dots).
Figure 8.
HOX genes are not dysregulated in LCLs of arhinia patients and muscle biopsies of FSHD2 patients. (A) Dot plots showing the FPKMs of the four clusters of HOX genes in LCLs of 10 arhinia patients (red) and 10 controls (black). (B) Dot plots showing the FPKMs of the four clusters of HOX genes in muscle biopsies of nine FSHD1 patients (blue), four FSHD2 patients carrying SMCHD1 mutations (red), two FSHD2 patients without SMCHD1 mutations (green), and eight controls (black).(C) A table listing other homeobox genes upregulated in Smchd1−/− mouse NPCs. (D) The top 20 list of the GO analysis of the 230 upregulated genes in Smchd1−/− mouse NPCs. GO terms that were not enriched with statistical significance were labeled with lighter colors. FPKM, fragments per kilobase of transcript per million mapped reads; FSHD2, facioscapulohumeral muscular dystrophy type 2; GO, gene ontology; ID, identifier; LCL; lymphoblastoid cell line; NPC, neural progenitor cell; SMCHD1, structural maintenance of chromosomes flexible hinge domain-containing 1.
We then investigated HOX expression in FSHD2 patients using RNA-seq data from muscle biopsies (n = 3). The HOX expression pattern was the same in controls and FSHD2 patients (Figure 8B, red dots). Muscle biopsies from FSHD1 patients (Figure 8B, blue dots) and FSHD2 patients without SMCHD1 mutations (Figure 8B, green dots) also exhibited similar HOX patterns. Together, we conclude that SMCHD1 mutations in the heterozygous state do not alter HOX expression in LCLs of arhinia patients and muscle biopsies of FSHD2 patients.
Derepression of genes involved in craniofacial development in Smchd1−/− mouse NPCs
Because it is not possible to study a homozygous SMCHD1 null in humans (see the Introduction), we turned again to the mouse to gain more insight into how the transcriptional program affected by SMCHD1 dysfunction might contribute to arhinia. We examined the function of other SMCHD1-regulated autosomal genes in mice. In addition to the Hox genes, Smchd1 ablation led to upregulation of five non-Hox homeobox genes (Figure 8C). Among them was Distal-less homeobox (Dlx) 2, a family member of the Dlx gene family that plays a crucial role in the development of cranial NCCs and surface ectoderm derivatives, such as olfactory and otic placodes (Panganiban and Rubenstein 2002). Mice carrying homozygous mutations of Dlx2 exhibit craniofacial defects, such as cleft palate (Qiu et al. 1995). Also identified was Ventral anterior homeobox 2 (Vax2), a critical regulator of eye development, with its inactivation in mice being associated with incomplete closure of the optic fissure and eye coloboma (Barbieri et al. 2002). Thus, dysfunction of SMCHD1-regulated developmental regulators causes anomalies overlapping with arhinia-associated malformations. Consistent with these observations, GO analyses of SMCHD1-repressed autosomal genes revealed the enrichment for GO terms associated with embryonic development, pattern specification, and morphogenesis (Figure 8D) (Table S7). On the other hand, a GO analysis of downregulated autosomal genes in Smchd1−/− mouse NPCs did not reveal statistically significant enrichment of any GO terms (Table S8). We conclude that SMCHD1 deficiency in mouse NPCs leads to a dysregulated transcriptional program, where genes critical for craniofacial development become aberrantly expressed. These findings are consistent with the possibility that SMCHD1 may contribute to the pathogenesis of the craniofacial anomalies in arhinia through the dysregulation of homeobox genes.
Discussion
Here, we have integrated RNA-seq and epigenomic profiling data of Smchd1−/− mouse cells to understand the molecular pathogenesis of two human disorders associated with SMCHD1 mutations: arhinia and FSHD2. Analysis of transcriptomic data from arhinia and FSHD2 patients demonstrate that the Xi chromosome remains dosage compensated in females. This finding is consistent with the absence of a sex bias in disease incidence or in clinical severity in patients with arhinia (Shaw et al. 2017 and N. Shaw, unpublished results). In fact, in FSHD multiplex families, females are more likely to be entirely asymptomatic or less symptomatic than male relatives (Zatz et al. 1995; Ricci et al. 1999, 2013; Tonini et al. 2004; Park et al. 2015) [however, this comparison may be confounded by the proposed protective effect of estrogen in FSHD, reviewed in Mul et al. (2018)]. Therefore, we conclude that XCI is not a contributor to the overall disease presentation and progression.
The absence of an effect on XCI may be somewhat surprising, but there are at least three possible explanations. First, all patients carry heterozygous mutations of SMCHD1 and thus have some amount of functional protein. While the residual SMCHD1 activity may be insufficient to support proper craniofacial development (arhinia) and maintenance of muscle health (FSHD2), subtotal reduction of SMCHD1 function does not appear to impact XCI. This differential sensitivity may be due to the fact that, as shown by both immunofluorescence and epigenomic analysis of SMCHD1-binding sites, SMCHD1 is more concentrated on the Xi than anywhere else in the genome (Blewitt et al. 2008; Wang et al. 2018). The Xi’s affinity for SMCHD1, as well as the multiple repressive mechanisms that synergize on the Xi, may render it less susceptible to subtotal reduction of normal SMCHD1 levels. Second, the missense SMCHD1 mutations associated with arhinia may be hypomorphs rather than null mutations. Indeed, SMCHD1 abundance does not appear to be affected in cells (Shaw et al. 2017). If they are hypomorphic mutations or if the missense affects a region of SMCHD1 that is not critical for XCI, dosage compensation in females could be preserved. Finally, given that the patients have different genetic backgrounds, there could be modifier genes that ameliorate the effect of SMCHD1 dysfunction. It is also possible that there is a transient X–A imbalance in human females during a very early developmental window, as transient X-linked CpG hypomethylation has been observed in female Smchd1MommeD1/+ mice at E8.5–10.5 (Blewitt et al. 2005, 2008). The patients could therefore represent survivors of delayed dosage compensation.
The resilience of X-linked genes to these mutations suggests that the necessity of intact SMCHD1 in silencing the Xi is different from susceptible loci on autosomes, such as DUX4 (Lemmers et al. 2012). Our study demonstrates clear effects of SMCHD1 deficiency on a subset of autosomal gene clusters. SMCHD1 participates in establishing not only the expression pattern, but also the epigenetic states and chromatin architectures of five autosomal gene clusters—Pcdha (Figure 5) and all four clusters of Hox genes (Figure 7, Figures S6, and S7)—in mice. Our finding is consistent with a SMCHD1 ChIP-seq study showing association of SMCHD1 with the Pcdh and Hox clusters in mouse neural stem cells (Chen et al. 2015), and a concurrent study implicating SMCHD1 in Hox regulation (Jansz et al. 2018a). At the chromatin level, Smchd1 ablation leads to enhanced binding of not only CTCF (Chen et al. 2015), but also cohesin at the Pcdha loci, coinciding with frequent enhancer–promoter contacts to facilitate transcription. At the Hox clusters, we observed erosion of H3K27me3 domains and decreased inter-subTAD interactions. These observations reinforce conclusions of previous work in which SMCHD1 was shown to weaken chromatin structures (TADs and compartments), enhance Polycomb recruitment, and repel the binding of canonical architectural proteins (Wang et al. 2018).
In support of a conserved role of human SMCHD1 in regulating PCDH and HOX, a previous study identified all three PCDH clusters, as well as the HOXB and HOXD clusters, to be CpG hypomethylated in peripheral blood mononuclear cells of non-FSHD-presenting female SMCHD1 mutation carriers. This study also reported slight upregulation (∼1.3–3-fold) of four PCDHB isoforms and one HOX gene in FSHD2 myoblast/myotube cultures, although the changes in PCDHB were also observed in FSHD1 cultures (Mason et al. 2017). Instead of PCDHB, we detected PCDHA dysregulation in the LCLs of two arhinia and the muscle biopsy of one FSHD2 patients. To our knowledge, PCDHA upregulation has not been reported in arhinia patients. Previous analysis of the same FSHD data sets reported PCDHB2 as an FSHD2-specific upregulated gene (Yao et al. 2014), whereas our analysis suggests that PCDHA/B derepression may be more specific to one FSHD2 patient. While these data support a link between SMCHD1 dysfunction and PCDHA misregulation, not all arhinia/FSHD2 patients exhibit PCDHA upregulation, even when carrying an identical SMCHD1 mutation (Figure 6B). The patterns of aberrant PCDHA upregulation observed in patients are also heterogeneous, sometimes accompanied by increased antisense transcription (Figure 6B). Together, these results suggest that regulation of the clustered PCDH genes is complex and may involve genetic or epigenetic modifiers that interact with SMCHD1. As the PCDHA cluster is a highly polymorphic region (Noonan et al. 2003) that is susceptible and tolerant to gene conversions (Miki et al. 2005) and deletions (Noonan et al. 2003) in humans, altered PCDHA expression may be related not to a change in SMCHD1 activity, but to one or more polymorphic SNPs in the PCDHA cluster in these patients. Another limitation of our study is that clustered PCDHs are lowly expressed in LCLs and myogenic cells, from which the patient RNA-seq data were generated, which likely explains why their upregulation does not reach statistical significance in DESeq analyses (Table S2). In light of the link between aberrant regulation of clustered PCDH genes and brain disorders (El Hajj et al. 2017), it may be relevant to examine PCDH expression in pertinent cell types (such as induced pluripotent stem cell-derived neurons) of these patients, and to evaluate potential neurological and psychiatric comorbidities (ClinialTrials.gov ID: NCT02032979).
However, we did not detect HOX gene dysregulation in arhinia and FSHD2 patients. While HOX genes may be more resilient to SMCHD1 dysfunction, it is also possible that SMCHD1 mutations are associated with ectopic or precocious HOX expression in other cell types, such as NCCs, or during an earlier developmental window. Indeed, although Hox genes remain repressed in Smchd1−/− neural stem cells derived from the forebrain of mouse embryos (Chen et al. 2015), posterior homeotic transformation of the axial skeleton has recently been reported for these animals (Jansz et al. 2018a). Furthermore, premature or delayed Hox expression also causes phenotypes, even when the final expression pattern is normal (Gérard et al. 1997; Zakany et al. 1997; Kondo and Duboule 1999; Juan and Ruddle 2003). In addition to Hox, Smchd1 ablation also leads to dysregulation of other homeobox genes critical for craniofacial development (Figure 8C). Additional work will be necessary to determine if these genes are dysregulated in the relevant cell types from patients, and their contribution to the pathogenesis of arhinia and FSHD2.
Acknowledgments
We thank S. Tapscott for providing genotypes of female FSHD2 patients; all members of the Lee laboratory for intellectual support; and M. Kuroda, S. Buratowski, and A. Gimelbrant for valuable advice during the thesis work of C.-Y.W. This work was supported by grants from the National Institutes of Health (NIH) (RO1 GM-090278 to J.T.L., K99 DE-026824 to H.B., and P01 GM-061354 to M.E.T.), the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01 ES-103315 and 1SI2 ES-025429-01 to N.D.S.), the Rett Syndrome Research Trust (J.T.L.), and the Howard Hughes Medical Institute (J.T.L.).
Author contributions: C.-Y.W. and J.T.L. conceived the project. C.-Y.W. designed, performed, and interpreted the analyses. H.B., N.D.S., and M.E.T. provided the arhinia patient RNA-seq data sets. C.-Y.W., N.D.S., and J.T.L. wrote the paper with input from all authors. J.T.L. is a cofounder of, and a scientific advisor for, Translate Bio and Fulcrum Therapeutics.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9633668.
Communicating editor: J. Schimenti
Literature Cited
- Anders S., and Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrey G., Montavon T., Mascrez B., Gonzalez F., Noordermeer D. et al. , 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167 10.1126/science.1234167 [DOI] [PubMed] [Google Scholar]
- Balaton B. P., and Brown C. J., 2016. Escape artists of the X chromosome. Trends Genet. 32: 348–359. 10.1016/j.tig.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Balaton B. P., Cotton A. M., and Brown C. J., 2015. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differ. 6: 35 10.1186/s13293-015-0053-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F., Roure V., Comet I., Leblanc B., Schuettengruber B. et al. , 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226. 10.1016/j.cell.2010.12.026 [DOI] [PubMed] [Google Scholar]
- Barbieri A. M., Broccoli V., Bovolenta P., Alfano G., Marchitiello A. et al. , 2002. Vax2 inactivation in mouse determines alteration of the eye dorsal-ventral axis, misrouting of the optic fibres and eye coloboma. Development 129: 805–813. [DOI] [PubMed] [Google Scholar]
- Berletch J. B., Yang F., Xu J., Carrel L., and Disteche C. M., 2011. Genes that escape from X inactivation. Hum. Genet. 130: 237–245. 10.1007/s00439-011-1011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch J. B., Ma W., Yang F., Shendure J., Noble W. S. et al. , 2015. Escape from X inactivation varies in mouse tissues. PLoS Genet. 11: e1005079 10.1371/journal.pgen.1005079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlivet S., Paquette D., Dumouchel A., Langlais D., Dostie J. et al. , 2013. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 9: e1004018 10.1371/journal.pgen.1004018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt M. E., Vickaryous N. K., Hemley S. J., Ashe A., Bruxner T. J. et al. , 2005. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl. Acad. Sci. USA 102: 7629–7634. 10.1073/pnas.0409375102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt M. E., Gendrel A. V., Pang Z., Sparrow D. B., Whitelaw N. et al. , 2008. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 40: 663–669. 10.1038/ng.142 [DOI] [PubMed] [Google Scholar]
- Bosma J. F., Henkin R. I., Christiansen R. L., and Herdt J. R., 1981. Hypoplasia of the nose and eyes, hyposmia, hypogeusia, and hypogonadotrophic hypogonadism in two males. J. Craniofac. Genet. Dev. Biol. 1: 153–184. [PubMed] [Google Scholar]
- Brideau N. J., Coker H., Gendrel A. V., Siebert C. A., Bezstarosti K. et al. , 2015. Independent mechanisms target SMCHD1 to trimethylated histone H3 lysine 9-modified chromatin and the inactive X chromosome. Mol. Cell. Biol. 35: 4053–4068. 10.1128/MCB.00432-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., Cooper P., Smith S. et al. , 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351: 329–331. 10.1038/351329a0 [DOI] [PubMed] [Google Scholar]
- Brown C. J., Ballabio A., Rupert J. L., Lafreniere R. G., Grompe M. et al. , 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44. 10.1038/349038a0 [DOI] [PubMed] [Google Scholar]
- Calabrese J. M., Sun W., Song L., Mugford J. W., Williams L. et al. , 2012. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151: 951–963. 10.1016/j.cell.2012.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrette L. L. G., Wang C. Y., Wei C., Press W., Ma W. et al. , 2018. A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc. Natl. Acad. Sci. USA 115: E668–E675. 10.1073/pnas.1715124115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Hu J., Moore D. L., Liu R., Kessans S. A. et al. , 2015. Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc. Natl. Acad. Sci. USA 112: E3535–E3544. 10.1073/pnas.1504232112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Dobson R. C., Lucet I. S., Young S. N., Pearce F. G. et al. , 2016. The epigenetic regulator Smchd1 contains a functional GHKL-type ATPase domain. Biochem. J. 473: 1733–1744. 10.1042/BCJ20160189 [DOI] [PubMed] [Google Scholar]
- Chen W. V., and Maniatis T., 2013. Clustered protocadherins. Development 140: 3297–3302. 10.1242/dev.090621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbellay F., and Duboule D., 2016. Topological domains, metagenes, and the emergence of pleiotropic regulations at Hox loci. Curr. Top. Dev. Biol. 116: 299–314. 10.1016/bs.ctdb.2015.11.022 [DOI] [PubMed] [Google Scholar]
- de Greef J. C., Krom Y. D., den Hamer B., Snider L., Hiramuki Y. et al. , 2018. Smchd1 haploinsufficiency exacerbates the phenotype of a transgenic FSHD1 mouse model. Hum. Mol. Genet. 27: 716–731. 10.1093/hmg/ddx437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C. M., 2012. Dosage compensation of the sex chromosomes. Annu. Rev. Genet. 46: 537–560. 10.1146/annurev-genet-110711-155454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y. et al. , 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D., 2007. The rise and fall of Hox gene clusters. Development 134: 2549–2560. 10.1242/dev.001065 [DOI] [PubMed] [Google Scholar]
- Duboule D., and Dolle P., 1989. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8: 1497–1505. 10.1002/j.1460-2075.1989.tb03534.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand N. C., Robinson J. T., Shamim M. S., Machol I., Mesirov J. P. et al. , 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3: 99–101. 10.1016/j.cels.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hajj N., Dittrich M., and Haaf T., 2017. Epigenetic dysregulation of protocadherins in human disease. Semin. Cell Dev. Biol. 69: 172–182. 10.1016/j.semcdb.2017.07.007 [DOI] [PubMed] [Google Scholar]
- Eskeland R., Leeb M., Grimes G. R., Kress C., Boyle S. et al. , 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38: 452–464. 10.1016/j.molcel.2010.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi S., Kakazu N., Taguchi Y., Hirayama T., Sasaki A. et al. , 2005. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet. 37: 171–176. 10.1038/ng1500 [DOI] [PubMed] [Google Scholar]
- Gdula M. R., Nesterova T. B., Pintacuda G., Godwin J., Zhan Y. et al. , 2019. The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat. Commun. 10: 30 10.1038/s41467-018-07907-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A. V., Apedaile A., Coker H., Termanis A., Zvetkova I. et al. , 2012. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell 23: 265–279. 10.1016/j.devcel.2012.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A. V., Tang Y. A., Suzuki M., Godwin J., Nesterova T. B. et al. , 2013. Epigenetic functions of smchd1 repress gene clusters on the inactive x chromosome and on autosomes. Mol. Cell. Biol. 33: 3150–3165. 10.1128/MCB.00145-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A. V., Attia M., Chen C. J., Diabangouaya P., Servant N. et al. , 2014. Developmental dynamics and disease potential of random monoallelic gene expression. Dev. Cell 28: 366–380. 10.1016/j.devcel.2014.01.016 [DOI] [PubMed] [Google Scholar]
- Gérard M., Zákány J., and Duboule D., 1997. Interspecies exchange of a Hoxd enhancer in vivo induces premature transcription and anterior shift of the sacrum. Dev. Biol. 190: 32–40. 10.1006/dbio.1997.8679 [DOI] [PubMed] [Google Scholar]
- Gordon C. T., Xue S., Yigit G., Filali H., Chen K. et al. , 2017. De novo mutations in SMCHD1 cause Bosma arhinia microphthalmia syndrome and abrogate nasal development. Nat. Genet. 49: 249–255. 10.1038/ng.3765 [DOI] [PubMed] [Google Scholar]
- Graham A., Papalopulu N., and Krumlauf R., 1989. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 57: 367–378. 10.1016/0092-8674(89)90912-4 [DOI] [PubMed] [Google Scholar]
- Guo Y., Monahan K., Wu H., Gertz J., Varley K. E. et al. , 2012. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl. Acad. Sci. USA 109: 21081–21086. 10.1073/pnas.1219280110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurzau A. D., Chen K., Xue S., Dai W., Lucet I. S. et al. , 2018. FSHD2- and BAMS-associated mutations confer opposing effects on SMCHD1 function. J. Biol. Chem. 293: 9841–9853. 10.1074/jbc.RA118.003104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C. et al. , 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., and Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Izpisúa-Belmonte J. C., Falkenstein H., Dollé P., Renucci A., and Duboule D., 1991. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 10: 2279–2289. 10.1002/j.1460-2075.1991.tb07764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansz N., Chen K., Murphy J. M., and Blewitt M. E., 2017. The epigenetic regulator SMCHD1 in development and disease. Trends Genet. 33: 233–243. 10.1016/j.tig.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Jansz N., Keniry A., Trussart M., Bildsoe H., Beck T. et al. , 2018a Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat. Struct. Mol. Biol. 25: 766–777. 10.1038/s41594-018-0111-z [DOI] [PubMed] [Google Scholar]
- Jansz N., Nesterova T., Keniry A., Iminitoff M., Hickey P. F. et al. , 2018b Smchd1 targeting to the inactive X is dependent on the Xist-HnrnpK-PRC1 pathway. Cell Rep. 25: 1912–1923.e9. 10.1016/j.celrep.2018.10.044 [DOI] [PubMed] [Google Scholar]
- Jones T. I., Chen J. C., Rahimov F., Homma S., Arashiro P. et al. , 2012. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 21: 4419–4430. 10.1093/hmg/dds284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan A. H., and Ruddle F. H., 2003. Enhancer timing of Hox gene expression: deletion of the endogenous Hoxc8 early enhancer. Development 130: 4823–4834. 10.1242/dev.00672 [DOI] [PubMed] [Google Scholar]
- Kehayova P., Monahan K., Chen W., and Maniatis T., 2011. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. USA 108: 17195–17200. 10.1073/pnas.1114357108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., and Duboule D., 1999. Breaking colinearity in the mouse HoxD complex. Cell 97: 407–417. 10.1016/S0092-8674(00)80749-7 [DOI] [PubMed] [Google Scholar]
- Krumlauf R., 1993. Hox genes and pattern formation in the branchial region of the vertebrate head. Trends Genet. 9: 106–112. 10.1016/0168-9525(93)90203-T [DOI] [PubMed] [Google Scholar]
- Kundu, S., F. Ji, H. Sunwoo, G. Jain, J. T. Lee et al., 2017 Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol. Cell 65: 432–446.e5 [corrigenda: Mol. Cell 71: 191 (2018)]. 10.1016/j.molcel.2017.01.009 [DOI] [PMC free article] [PubMed]
- Lee J. T., 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 12: 815–826. 10.1038/nrm3231 [DOI] [PubMed] [Google Scholar]
- Lemmers R. J., Tawil R., Petek L. M., Balog J., Block G. J. et al. , 2012. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 44: 1370–1374. 10.1038/ng.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers R. J., Goeman J. J., van der Vliet P. J., van Nieuwenhuizen M. P., Balog J. et al. , 2015. Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet. 24: 659–669. 10.1093/hmg/ddu486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong H. S., Chen K., Hu Y., Lee S., Corbin J. et al. , 2013. Epigenetic regulator Smchd1 functions as a tumor suppressor. Cancer Res. 73: 1591–1599. 10.1158/0008-5472.CAN-12-3019 [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Li S. M., Valo Z., Wang J., Gao H., Bowers C. W. et al. , 2012. Transcriptome-wide survey of mouse CNS-derived cells reveals monoallelic expression within novel gene families. PLoS One 7: e31751 10.1371/journal.pone.0031751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes A. M., Arnold-Croop S. E., Amorim A., and Carrel L., 2011. Clustered transcripts that escape X inactivation at mouse XqD. Mamm. Genome 22: 572–582. 10.1007/s00335-011-9350-6 [DOI] [PubMed] [Google Scholar]
- Luijk R., Wu H., Ward-Caviness C. K., Hannon E., Carnero-Montoro E. et al. , 2018. Autosomal genetic variation is associated with DNA methylation in regions variably escaping X-chromosome inactivation. Nat. Commun. 9: 3738 10.1038/s41467-018-05714-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y., Panning B., Dausman J., Strauss W., and Jaenisch R., 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11: 156–166. 10.1101/gad.11.2.156 [DOI] [PubMed] [Google Scholar]
- Marks H., Kerstens H. H., Barakat T. S., Splinter E., Dirks R. A. et al. , 2015. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 16: 149 [corrigenda: Genome Biol. 17: 22 (2016)] 10.1186/s13059-015-0698-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason A. G., Slieker R. C., Balog J., Lemmers R., Wong C. J. et al. , 2017. SMCHD1 regulates a limited set of gene clusters on autosomal chromosomes. Skelet. Muscle 7: 12 10.1186/s13395-017-0129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki R., Hattori K., Taguchi Y., Tada M. N., Isosaka T. et al. , 2005. Identification and characterization of coding single-nucleotide polymorphisms within human protocadherin-alpha and -beta gene clusters. Gene 349: 1–14. 10.1016/j.gene.2004.11.044 [DOI] [PubMed] [Google Scholar]
- Mira-Bontenbal H., and Gribnau J., 2016. New Xist-interacting proteins in X–chromosome inactivation. Curr. Biol. 26: R338–R342. 10.1016/j.cub.2016.05.009 [DOI] [PubMed] [Google Scholar]
- Monahan K., Rudnick N. D., Kehayova P. D., Pauli F., Newberry K. M. et al. , 2012. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proc. Natl. Acad. Sci. USA 109: 9125–9130. 10.1073/pnas.1205074109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould A. W., Pang Z., Pakusch M., Tonks I. D., Stark M. et al. , 2013. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin 6: 19 10.1186/1756-8935-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountoufaris G., Chen W. V., Hirabayashi Y., O’Keeffe S., Chevee M. et al. , 2017. Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science 356: 411–414. 10.1126/science.aai8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mul K., Horlings C. G. C., Voermans N. C., Schreuder T. H. A., and van Engelen B. G. M., 2018. Lifetime endogenous estrogen exposure and disease severity in female patients with facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 28: 508–511. 10.1016/j.nmd.2018.02.012 [DOI] [PubMed] [Google Scholar]
- Noonan J. P., Li J., Nguyen L., Caoile C., Dickson M. et al. , 2003. Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am. J. Hum. Genet. 72: 621–635. 10.1086/368060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E. P., Lajoie B. R., Schulz E. G., Giorgetti L., Okamoto I. et al. , 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa R. S., Nagao K., Igami K. T., Shibata S., Shirai N. et al. , 2013. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat. Struct. Mol. Biol. 20: 566–573. 10.1038/nsmb.2532 [DOI] [PubMed] [Google Scholar]
- Panganiban G., and Rubenstein J. L., 2002. Developmental functions of the Distal-less/Dlx homeobox genes. Development 129: 4371–4386. [DOI] [PubMed] [Google Scholar]
- Park H. J., Hong J. M., Lee J. H., Lee H. S., Shin H. Y. et al. , 2015. Low D4Z4 copy number and gender difference in Korean patients with facioscapulohumeral muscular dystrophy type 1. Neuromuscul. Disord. 25: 859–864. 10.1016/j.nmd.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Parker H. J., Pushel I., and Krumlauf R., 2018. Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol. 444: S67–S78. 10.1016/j.ydbio.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Pinter S. F., Sadreyev R. I., Yildirim E., Jeon Y., Ohsumi T. K. et al. , 2012. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 22: 1864–1876. 10.1101/gr.133751.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M., Bulfone A., Martinez S., Meneses J. J., Shimamura K. et al. , 1995. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 9: 2523–2538. 10.1101/gad.9.20.2523 [DOI] [PubMed] [Google Scholar]
- Ribich S., Tasic B., and Maniatis T., 2006. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. USA 103: 19719–19724. 10.1073/pnas.0609445104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci E., Galluzzi G., Deidda G., Cacurri S., Colantoni L. et al. , 1999. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann. Neurol. 45: 751–757. [DOI] [PubMed] [Google Scholar]
- Ricci G., Scionti I., Sera F., Govi M., D’Amico R. et al. , 2013. Large scale genotype-phenotype analyses indicate that novel prognostic tools are required for families with facioscapulohumeral muscular dystrophy. Brain 136: 3408–3417. 10.1093/brain/awt226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacconi S., Lemmers R. J., Balog J., van der Vliet P. J., Lahaut P. et al. , 2013. The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am. J. Hum. Genet. 93: 744–751. 10.1016/j.ajhg.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y., Nagao K., Blewitt M., Sasaki H., Obuse C. et al. , 2018. Role of SmcHD1 in establishment of epigenetic states required for the maintenance of the X-inactivated state in mice. Development 145: dev166462. 10.1242/dev.166462 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S., Sugar R., Dimond A., Javierre B. M., Armstrong H. et al. , 2015. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 47: 1179–1186. 10.1038/ng.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., and Pirrotta V., 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8: 9–22. 10.1038/nrg1981 [DOI] [PubMed] [Google Scholar]
- Shaw N. D., Brand H., Kupchinsky Z. A., Bengani H., Plummer L. et al. , 2017. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 49: 238–248 (erratum: Nat. Genet. 49: 969). 10.1038/ng.3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N., and Duboule D., 2009. Epigenetic temporal control of mouse Hox genes in vivo. Science 324: 1320–1323. 10.1126/science.1171468 [DOI] [PubMed] [Google Scholar]
- Splinter E., de Wit E., Nora E. P., Klous P., van de Werken H. J. et al. , 2011. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 25: 1371–1383. 10.1101/gad.633311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starmer J., and Magnuson T., 2009. A new model for random X chromosome inactivation. Development 136: 1–10. 10.1242/dev.025908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A., and Waxman D. J., 2013. Genome-wide analysis of chromatin states reveals distinct mechanisms of sex-dependent gene regulation in male and female mouse liver. Mol. Cell. Biol. 33: 3594–3610. 10.1128/MCB.00280-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface L. E., Thornton S. R., and Boyer L. A., 2010. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 7: 288–298. 10.1016/j.stem.2010.08.004 [DOI] [PubMed] [Google Scholar]
- Tasic B., Nabholz C. E., Baldwin K. K., Kim Y., Rueckert E. H. et al. , 2002. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10: 21–33. 10.1016/S1097-2765(02)00578-6 [DOI] [PubMed] [Google Scholar]
- Tonini M. M., Passos-Bueno M. R., Cerqueira A., Matioli S. R., Pavanello R. et al. , 2004. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD). Neuromuscul. Disord. 14: 33–38. 10.1016/j.nmd.2003.07.001 [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Jégu T., Chu H. P., Oh H. J. and Lee J. T., 2018. SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 174: 406–421.e25. 10.1016/j.cell.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Y., Colognori D., Sunwoo H., Wang D., and Lee J. T., 2019. PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat. Commun. 10: 2950 10.1038/s41467-019-10755-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Su H., and Bradley A., 2002. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 16: 1890–1905. 10.1101/gad.1004802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie A. O., 2017. Many faces of SMCHD1. Nat. Genet. 49: 176–178. 10.1038/ng.3776 [DOI] [PubMed] [Google Scholar]
- Wu Q., and Maniatis T., 1999. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97: 779–790. 10.1016/S0092-8674(00)80789-8 [DOI] [PubMed] [Google Scholar]
- Wu Q., Zhang T., Cheng J. F., Kim Y., Grimwood J. et al. , 2001. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 11: 389–404. 10.1101/gr.167301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A., 2011. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 12: 542–553. 10.1038/nrg3035 [DOI] [PubMed] [Google Scholar]
- Yagi T., 2013. Genetic basis of neuronal individuality in the mammalian brain. J. Neurogenet. 27: 97–105. 10.3109/01677063.2013.801969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Babak T., Shendure J., and Disteche C. M., 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 20: 614–622. 10.1101/gr.103200.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Kirby J. E., Sunwoo H., and Lee J. T., 2016. Female mice lacking Xist RNA show partial dosage compensation and survive to term. Genes Dev. 30: 1747–1760. 10.1101/gad.281162.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Snider L., Balog J., Lemmers R. J., Van Der Maarel S. M. et al. , 2014. DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet. 23: 5342–5352. 10.1093/hmg/ddu251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J., Gerard M., Favier B., and Duboule D., 1997. Deletion of a HoxD enhancer induces transcriptional heterochrony leading to transposition of the sacrum. EMBO J. 16: 4393–4402. 10.1093/emboj/16.14.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatz M., Marie S. K., Passos-Bueno M. R., Vainzof M., Campiotto S. et al. , 1995. High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy families. Am. J. Hum. Genet. 56: 99–105. [PMC free article] [PubMed] [Google Scholar]
- Zipursky S. L., and Sanes J. R., 2010. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell 143: 343–353. 10.1016/j.cell.2010.10.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All next-generation sequencing data are available at the GEO with the accession numbers GSE54016, GSE48109, GSE65747, GSE99991, GSE56787, and GSE126257. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9633668.