Table 3. Fractional coordinates and isotropic displacement parameters (Å2) for LiMHC6H5O7 from XRD/DFT calculation and NPD on POWGEN.
The first row in each entry corresponds to the XRD/DFT calculation, the second corresponds to NPD on POWGEN, and the third row is the difference between the X-ray/DFT and NPD values. The unit-cell parameters and goodness of fit are also provided in the header of the table. Same-atom species are constrained to have the same thermal parameters (U
iso). The position of Li is fixed for the refinement.Space group 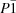 . Lattice parameters: a = 6.2844 (6), b = 6.3148 (4), c = 10.8249 (17) Å (DFT), a = 6.2848 (4), b = 6.3142 (3), c = 10.8259 (10) Å (NPD), α = 93.059 (4), β = 102.951 (3), γ = 96.094 (2)° (DFT), α = 93.049 (3), β = 102.960 (2), γ = 96.093 (1)° (NPD). Goodness-of-fit parameters: R
wp = 2.56% (DFT); R
wp = 1.57 (NPD).
. Lattice parameters: a = 6.2844 (6), b = 6.3148 (4), c = 10.8249 (17) Å (DFT), a = 6.2848 (4), b = 6.3142 (3), c = 10.8259 (10) Å (NPD), α = 93.059 (4), β = 102.951 (3), γ = 96.094 (2)° (DFT), α = 93.049 (3), β = 102.960 (2), γ = 96.093 (1)° (NPD). Goodness-of-fit parameters: R
wp = 2.56% (DFT); R
wp = 1.57 (NPD).
| Atom | x | y | z | U iso (Å2) |
|---|---|---|---|---|
| C1 | 1.1297 | 0.5120 | 0.3030 | 0.0099 (8) |
| 1.1334 (12) | 0.4972 (14) | 0.2932 (8) | 0.0096 (7) | |
| 0.0037 | 0.0148 | 0.0098 | 0.0003 | |
| C2 | 1.0220 | 0.6921 | 0.3505 | 0.0099 (8) |
| 1.0307 (13) | 0.6930 (12) | 0.3520 (8) | 0.0096 (7) | |
| 0.0087 | 0.0009 | 0.0015 | 0.0003 | |
| C3 | 0.8044 | 0.7464 | 0.2671 | 0.0099 (8) |
| 0.8230 (12) | 0.7555 (13) | 0.2666 (7) | 0.0096 (7) | |
| 0.0186 | 0.0091 | 0.0004 | 0.0003 | |
| C4 | 0.7260 | 0.9129 | 0.3512 | 0.0099 (8) |
| 0.7477 (12) | 0.9280 (13) | 0.3467 (8) | 0.0096 (7) | |
| 0.0217 | 0.0151 | 0.0046 | 0.0003 | |
| C5 | 0.5593 | 1.0569 | 0.2916 | 0.0099 (8) |
| 0.5612 (12) | 1.0602 (12) | 0.2852 (8) | 0.0096 (7) | |
| 0.0019 | 0.0032 | 0.0064 | 0.0003 | |
| C6 | 0.8475 | 0.8439 | 0.1456 | 0.0099 (8) |
| 0.8486 (12) | 0.8225 (10) | 0.1407 (6) | 0.0096 (7) | |
| 0.0011 | 0.0214 | 0.0049 | 0.0003 | |
| H7 | 1.1441 | 0.8338 | 0.3645 | 0.086 (4) |
| 1.1556 (32) | 0.8050 (30) | 0.3599 (17) | 0.071 (4) | |
| 0.0115 | 0.0288 | 0.0046 | 0.0150 | |
| H8 | 0.9965 | 0.6587 | 0.4444 | 0.086 (2) |
| 1.0319 (32) | 0.6457 (32) | 0.4529 (20) | 0.071 (4) | |
| 0.0354 | 0.0129 | 0.0084 | 0.0150 | |
| H9 | 0.8683 | 1.0210 | 0.4035 | 0.086 (4) |
| 0.8864 (28) | 1.0670 (31) | 0.3929 (16) | 0.071 (4) | |
| 0.0181 | 0.0459 | 0.0105 | 0.0150 | |
| H10 | 0.6582 | 0.8308 | 0.4232 | 0.086 (4) |
| 0.6962 (32) | 0.8455 (30) | 0.4299 (19) | 0.071 (4) | |
| 0.0380 | 0.0146 | 0.0066 | 0.0150 | |
| O11 | 1.2272 | 0.4066 | 0.3961 | 0.043 (2) |
| 1.2213 (17) | 0.4122 (16) | 0.3821 (10) | 0.033 (1) | |
| 0.0059 | 0.0056 | 0.0140 | 0.0100 | |
| O12 | 1.1355 | 0.4734 | 0.1906 | 0.043 (2) |
| 1.1159 (14) | 0.4687 (16) | 0.1853 (10) | 0.033 (1) | |
| 0.0196 | 0.0048 | 0.0053 | 0.0100 | |
| O13 | 0.5363 | 1.2099 | 0.3681 | 0.043 (2) |
| 0.5219 (15) | 1.1799 (17) | 0.3724 (9) | 0.033 (1) | |
| 0.0144 | 0.0300 | 0.0043 | 0.0100 | |
| O14 | 0.4561 | 1.0249 | 0.1774 | 0.043 (2) |
| 0.4896 (15) | 1.0413 (15) | 0.1760 (9) | 0.033 (1) | |
| 0.0335 | 0.0164 | 0.0013 | 0.0100 | |
| O15 | 0.9607 | 1.0242 | 0.1623 | 0.043 (2) |
| 0.9879 (14) | 0.9871 (15) | 0.9871 (15) | 0.033 (1) | |
| 0.0272 | 0.0371 | 0.0132 | 0.0100 | |
| O16 | 0.7692 | 0.7394 | 0.0397 | 0.043 (2) |
| 0.7653 (14) | 0.7312 (17) | 0.0361 (9) | 0.033 (1) | |
| 0.0039 | 0.0082 | 0.0036 | 0.0100 | |
| O17 | 0.6482 | 0.5615 | 0.2282 | 0.043 (2) |
| 0.6520 (15) | 0.5738 (17) | 0.2382 (10) | 0.033 (1) | |
| 0.0038 | 0.0123 | 0.0101 | 0.0100 | |
| H18 | 0.6337 | 0.4792 | 0.3003 | 0.086 (4) |
| 0.6306 (36) | 0.5102 (35) | 0.2968 (17) | 0.071 (4) | |
| 0.0031 | 0.0310 | 0.0035 | 0.0150 | |
| K19 | 0.2930 | 0.6951 | −0.0061 | 0.081 (8) |
| 0.2920 (31) | 0.7146 (30) | −0.0009 (16) | 0.056 (6) | |
| 0.0010 | 0.0195 | 0.0052 | 0.0052 | |
| Li20 | 0.7930 | 0.8114 | 0.8742 | 0.6 |
| 0.27 (4) | ||||
| H21 | 1.3404 | 0.3134 | 0.3710 | 0.086 (4) |
| 1.3641 (33) | 0.2933 (34) | 0.3777 (17) | 0.071 (4) | |
| 0.0238 | 0.0202 | 0.0067 | 0.0150 |
