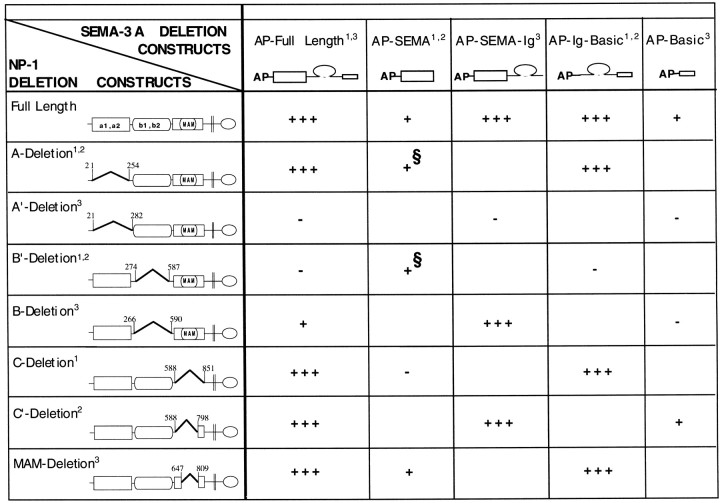
Table 1.
Summary of the domain mapping of SEMA-3A binding to neuropilin-1
A compendium of the results reported for the binding of SEMA-3A to neuropilin-1 as described: 1this paper;2Giger et al., 1998b; and 3Nakamura et al., 1998. Full-length and partial AP-tagged SEMA-3A constructs (top row) were tested for binding to various neuropilin-1 constructs (first column). Schematics of neuropilin-1 deletion constructs from different laboratories are shown with the amino acid numbers used to define domain deletion boundaries. Neuropilin-1 deletion constructs with break points differing by 12 amino acids or less are grouped together. Binding results are expressed as follows: +++, strong binding; +, weak binding; −, no detectable binding; and ( ), not tested. §, Results that differed between two laboratories in experiments using similar reagents. Giger et al. (1998b) observed no binding, whereas we detected binding after developing the reactions for very long times.