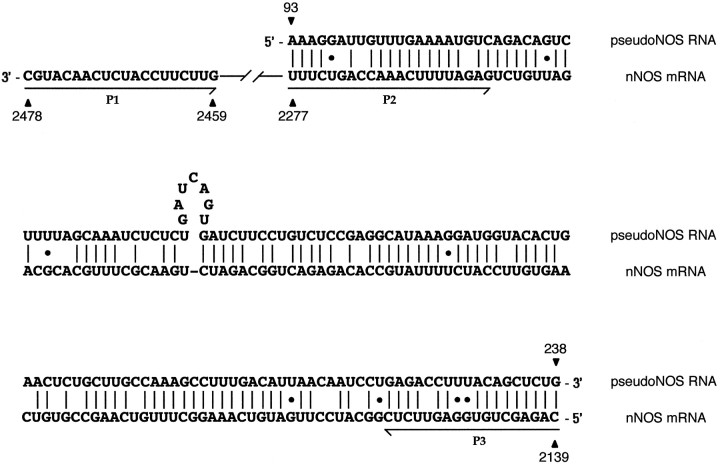
Fig. 2.
Alignment of the antisense region (93–238 nt) of the pseudo-NOS transcript with its complementary counterpart in the nNOS mRNA. In this alignment, there is ∼80% complementarity. The non-Watson–Crick G–U base pairs that are common in RNA secondary structure are shown by dots. The positions of three primers used in the identification of RNA–RNA duplexes areunderlined and named. Primers in positions expected to be protected from ribonuclease A (within the proposed duplex) are P2 and P3. The primer located outside of the protected area is called P1. Further explanation of the ribonuclease A protection experiment is provided in Figure 5. Details of procedures are in Materials and Methods.