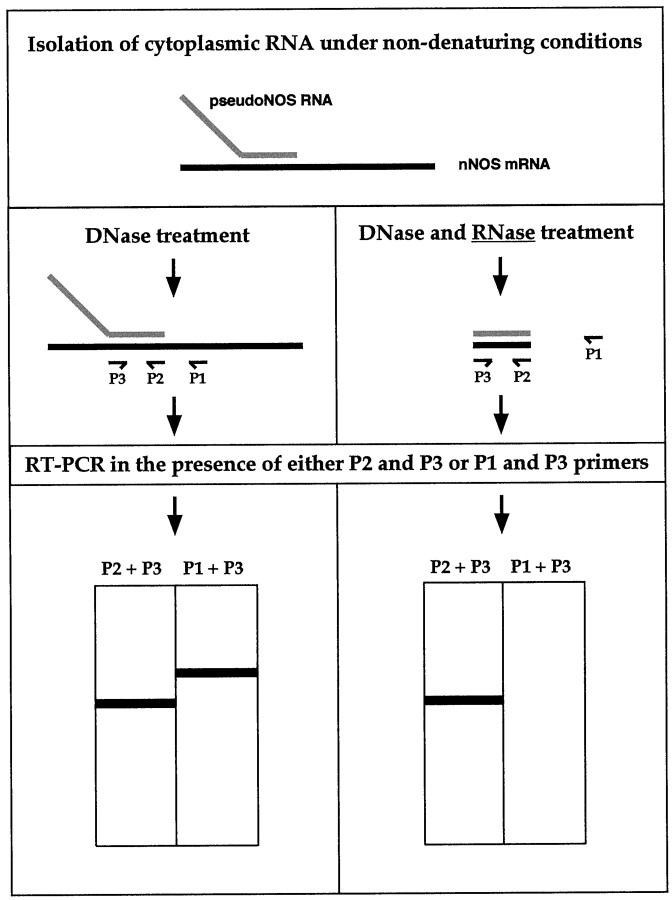
Fig. 5.
A schematic diagram showing the major steps of the ribonuclease protection procedure used to detect RNA–RNA duplexesin vivo. To preserve possible RNA–RNA hybrids, cytoplasmic RNA was purified from the CNS under nondenaturing conditions. To identify our hypothesized RNA–RNA duplex, the RNA has to be treated with RNase A, an enzyme that cleaves single-stranded but not double-stranded RNA molecules. After RNase A treatment, reverse transcription reactions in the presence of either P2 primer (located within the protected area) or P1 primer (located outside the protected area) are performed. After adding the P3 primer, a cDNA generated in the first reaction could be amplified using PCR and then will be revealed as a single band of the expected size by electrophoresis. In contrast, no cDNA could be produced in the second reverse transcription reaction, and subsequently, no PCR product is expected. In theleft column, the predicted results of the control experiments (no RNase A treatment) are summarized. Two RT-PCR products should be detected in the reverse transcription reaction: one generated by P2 and P3 and the other by P1 and P3.