Abstract
We have identified a mammalian opsin, encephalopsin, that shows strong and specific expression in the brain. Encephalopsin defines a new family of opsins and shows highest homology to vertebrate retinal and pineal opsins. Encephalopsin is highly expressed in the preoptic area and paraventricular nucleus of the hypothalamus, both regions implicated in encephalic photoreception in nonmammalian vertebrates. In addition, encephalopsin shows highly patterned expression in other regions of the brain, being enriched in selected regions of the cerebral cortex, cerebellar Purkinje cells, a subset of striatal neurons, selected thalamic nuclei, and a subset of interneurons in the ventral horn of the spinal cord. Rostrocaudal gradients of encephalopsin expression are present in the cortex, cerebellum, and striatum. Radial stripes of encephalopsin expression are seen in the cerebellum. In the cortex and cerebellum, encephalopsin expression is considerably higher and more highly patterned in the adult than in the neonate. Encephalopsin is the first putative extraocular opsin identified in mammals and may play a role in encephalic photoreception.
Keywords: photoreceptor, circadian, photopigment, pineal, retina, in situ hybridization, Purkinje, stripes, cerebellum, hypothalamus, deep brain
Vertebrate phototransduction serves multiple purposes. Visual phototransduction has received the great bulk of attention and has been extensively characterized at a molecular level (Palczewski, 1994). The molecular mechanism of nonvisual phototransduction, geared toward detecting changes in ambient light intensity and entrainment of circadian and other biological rhythms, has been generally much less well studied. Although retinal photoreceptors mediate visual phototransduction, the identity of the photoreceptors mediating circadian photoentrainment is unknown (Foster, 1998; Green, 1998). The photoreceptors mediating circadian photoreception may include both the rods and cones of the retina and other as yet unidentified sites (Provencio and Foster, 1995; Menaker et al., 1997). Cryptochromes, homologs of the blue-light receptor from plants, have been shown recently to play a role in light entrainment of circadian rhythms in both mammals and Drosophila (Thresher et al., 1998). However, it is not yet clear whether they are actual photoreceptors or part of the molecular machinery coupling phototransduction to the circadian clock.
Although visual phototransduction is specifically retinal, nonvisual phototransduction occurs in many extraretinal sites (Menaker et al., 1997). These include the pineal complex, the brain, the iris, melanocytes of the skin, and possibly other sites. Although extraretinal phototransduction has been extensively studied in nonmammalian vertebrates, adult mammals have traditionally been thought to lack extraretinal photoreceptors (Foster, 1998), given the fact that intact eyes are needed to entrain adult mammalian circadian behavioral rhythms. Recent work, however, has raised the possibility that mammals may indeed possess extraretinal photoreceptors. In humans there is evidence of circadian entrainment mediated by light applied to the backs of the knees (Campbell and Murphy, 1998). Moreover, free-running circadian rhythms have been identified in a variety of cultured mammalian cells (Balsalobre et al., 1998) and tissues (Zylka et al., 1998), although it is not yet clear whether light can directly entrain rhythms at these sites.
The molecular identity of the photopigments involved in nonvisual phototransduction is not fully known. The past few years have seen the identification of a variety of novel opsins in nonmammalian vertebrates, including the pineal complex-specific pinopsin (Okano et al., 1994; Max et al., 1995) and parapinopsin (Blackshaw and Snyder, 1997b), melanopsin, which is expressed in melanophores and retinal horizontal cells (Provencio et al., 1998), and vertebrate ancient (VA) opsin, which is expressed in retinal horizontal and amacrine cells (Soni et al., 1998). The molecular characteristics of the encephalic photoreceptor system are unknown, although in amphibians and birds (Provencio et al., 1998; Wada et al., 1998; Yoshikawa et al., 1998) expression of either rhodopsin, pinopsin, or melanopsin has been reported in the diencephalon, depending on the species examined. Molecular evidence of extraretinal photoreceptors in mammals has been lacking, however, except for the expression of visual opsins in the pineal gland (Blackshaw and Snyder, 1997a) and cryptochromes, which are expressed in a wide range of neuronal and peripheral tissues (Miyamoto and Sancar, 1998).
We now describe the identification, cloning, and characterization of a novel mammalian opsin, encephalopsin, which is highly concentrated in the brain and testes and localized to discrete areas of the brain.
MATERIALS AND METHODS
Cloning of encephalopsin. A 180 bp fragment of a novel opsin from Xenopus laevis covering transmembrane domains 6 and 7 (S. Blackshaw and S. H. Snyder, unpublished observations) was used to screen the database of expressed sequence tags (dbESTs) using TBLASTX. A homologous clone from mouse (GenBank accession number AA276584) was identified. This was then used as a query to search the dbESTs, and several highly homologous human ESTs (GenBank accession numbers T83438, AA088574, AA367654, R70620, andR76620) were identified. After the completion of one round of 5′ rapid amplification of cDNA ends (RACE), which extended the mouse encephalopsin cDNA to 335 bp from the start of the coding sequence, another human EST (accession number AA297478) was identified that extended as far as the third transmembrane domain of the encephalopsin-coding sequence.
These sequences were used to design 5′-RACE primers for amplification of full-length cDNAs (Frohman et al., 1988). Human and mouse cerebellar and testes RNA was used as first-strand cDNA for RACE, with GC-MELT (Clontech, Palo Alto, CA) added to the final round of RACE to obtain the GC-rich 5′ end. Human RNA was purchased from Clontech, whereas mouse total RNA was isolated via RNeasy (Qiagen, Hilden, Germany). Three successive rounds of 5′ RACE were performed to obtain the mouse 5′ end, whereas two rounds were required to obtain the human 5′ end. DNA sequence was confirmed by sequencing multiple independent cDNAs and via genomic sequencing, which was also used to confirm the position of intron–exon junctions. 5′-RACE primers for mouse encephalopsin covered residues 261–285, 471–498, and 1180–1207, as numbered from the start of the cDNA-coding sequence. Human 5′-RACE primers covered residues 241–260 and 525–554. 3′ sequences for mouse and human encephalopsin were obtained via sequencing multiple partial-length ESTs and, in the mouse, via genomic DNA sequencing.
Genomic clones for mouse encephalopsin were isolated via a combination of genomic library screening and inverse PCR. A RACE product corresponding to residues 348–1207 of the mouse encephalopsin-coding sequence was used as a probe in genomic DNA library screening and was used to screen 800,000 individual plaques. Two overlapping clones were isolated that covered an interval that included the interval in the encephalopsin-coding sequence from exon 2 to the end of the coding sequence. Exon 1 and the flanking genomic sequence were isolated via inverse PCR (Pang and Knecht, 1997). Multiple clones were recovered by inverse PCR, and their sequences checked against the clones obtained by 5′ RACE. Full-length mouse and human encephalopsin cDNAs were obtained by reverse transcription (RT)-PCR using proofreading Pfu polymerase from cerebellar cDNA with primers directed against the start and end of the coding sequences.
Phylogenetic analysis. Phylogenetic analysis was performed with the CLUSTALW function of Macvector 6.5. Swiss-Protein accession numbers of the sequences used to calculate the tree are as follows: human rhodopsin, P08100; human blue cone pigment, P03999; human green cone pigment, P04001; human red cone pigment, P04000; goldfish UV cone pigment, Q90309; chick pinopsin, P51475; salmon VA opsin, O13018; catfish parapinopsin, O42266; mouse peropsin, O35214; human peropsin, O14718; bovine RGR, P47803; human RGR, P47804; squid retinochrome, P23820; squid rhodopsin, P31356; octopus rhodopsin,P09241; Drosophila Rh1 opsin, P06002; DrosophilaRh2 opsin, P08099; and Xenopus melanopsin, AF014797.
Northern blotting. Northern blotting was performed using a random-primed probe corresponding to residues 348–1207 of mouse encephalopsin. Twenty micrograms of total RNA were loaded per lane. RNA was isolated via RNeasy (Qiagen) and was confirmed to be of high quality by ethidium bromide staining of the gel before transfer. The blot was exposed to Kodak MR film (Eastman Kodak, Rochester, NY) for 1 week. Hybridization to a β-actin probe demonstrated equal loading in each lane (data not shown).
In situ hybridization. cRNA probes corresponding to residues 348–1207 of mouse encephalopsin were used for both radioactive and nonradioactive in situ hybridization. Sense control probes were tested in parallel against each time point and tissue and at no point showed specific hybridization. Digoxygeninin situ hybridization was performed on fresh-frozen sections as described previously (Blackshaw and Snyder, 1997). Color reactions were allowed to proceed for 5 d. C57B6 mice were used in developmental studies. Each developmental time point was run in parallel with multiple slides for each. For examination of pigmented tissues, albino CD-1 mice were also used. No significant strain differences in expression were observed. Radioactive in situhybridization was performed essentially as described elsewhere (Vassar et al., 1994), except that 107 cpm of33P-labeled sense and antisense probe was used per slide. Slides were exposed to Kodak MR-2 film for 4 d.
RESULTS
Molecular cloning of encephalopsin
In the course of previous work, we have identified several novel putative extraretinal opsins from cold-blooded vertebrates via PCR with primers designed to regions of homology shared by pinopsin and parapinopsin but not retinal opsins (Blackshaw and Snyder, unpublished observations). We used one of these novel partial-length cDNAs isolated from Xenopus as a query in a search for related genes in the dbESTs. We identified novel transcripts from human and mouse with high homology to a variety of retinal and extra retinal opsins, although they did not prove to be true homologs of the novelXenopus opsin used as a query sequence.
To clone encephalopsin, we obtained full-length cDNAs by RACE from human and mouse brain. The 3′-end sequence was obtained by sequencing multiple independent ESTs. Full-length clones were reconstructed by PCR. The mouse and human sequences have been deposited in GenBank under accession numbers AF140241 and AF140242, respectively.
Analysis of the human and mouse encephalopsin sequences reveals high homology to other members of the opsin family (Fig.1). A number of molecular features establish that encephalopsin fulfills the criteria expected of a functional opsin. Such features include the following:
Fig. 1.

Primary sequence of human and mouse encephalopsin. Sequences are aligned via CLUSTALW with members of each of the main classes of vertebrate retinal and extraretinal opsins. Residues conserved in the majority of sequences are in darkly shaded boxes, whereas similar residues are in lightly shaded boxes. The position of the lysine in the seventh transmembrane domain that forms a Schiff base with retinal is indicated by anasterisk, whereas the position of the counterion in the third transmembrane domain is indicated by a diamond. Positions of the introns present in encephalopsin and the rod, cone, and pineal opsins are indicated with a black arrowhead. The gray arrowhead indicates the intron that is present in rod and cone opsins, along with pinopsin, but is absent in encephalopsin and parapinopsin. The white arrowheadindicates the position of the N-terminal intron found in the red- and green-sensitive opsins.
(1) Encephalopsin possesses a lysine at position 297 in mouse encephalopsin (296 in bovine rhodopsin), the same as in all known functional opsins (Schertler, 1998). This lysine is required to form a Schiff base with 11-cis-retinal. This lysine occurs only in opsins and in retinaldehyde-binding proteins such as retinochrome (Hara-Nishimura et al., 1990) and the RGR protein from retinal pigmented epithelium (Shen et al., 1994).
(2) In all functional opsins the positive charge created by the Schiff base formed by lysine and retinal is neutralized by an amino acid in the third transmembrane domain (Schertler, 1998). In all vertebrate functional opsins this counterion is an acidic residue (Sakmar et al., 1989; Nathans, 1990), whereas in invertebrates an aromatic amino acid, usually tryptophan or tyrosine, is found at this site. At position 115 (corresponding to 113 in bovine rhodopsin), encephalopsin possesses an aspartate, consistent with a functional role as an opsin. Although the acidic counterion in retinal and pineal opsins is typically a glutamate, aspartate can fully substitute in this role, with the only effect being a slight red shift in maximum absorbance wavelength (Sakmar et al., 1989).
(3) Vertebrate retinal and pineal opsins possess a common intron–exon junction organization (Nathans et al., 1986; Max et al., 1995;Yokoyama, 1996). Encephalopsin displays this pattern. These junctions are at nucleotides 367, 687, and 936 of the coding sequence (Fig. 1). These are identical to the positions of the intron–exon junctions of parapinopsin (Blackshaw and Snyder, 1997b). The intron typically found at the end of the fourth transmembrane domain, conserved in the retinal opsins and pinopsin, and the intron found in the N-terminal domain of long-wave opsins are both absent.
(4) Many of the residues of the transmembrane domains identified as involved in spectral tuning in vertebrate opsins are conserved in encephalopsin. These include Gly-92, Gly-119, Ser-163, Phe-262, and Ala-296 (Neitz et al., 1991; Merbs and Nathans, 1992; Lin et al., 1998). These correspond to residues 90, 119, 164, 261, and 295, respectively, in bovine rhodopsin.
(5) Opsins typically contain high densities of serine and threonine residues in the C-terminal region, which are substrates for phosphorylation by various receptor kinases (Schertler, 1998). Encephalopsin similarly displays enrichment in serine and threonine in the C-terminal area.
(6) Opsins usually incorporate a site for N-linked glycosylation in the N terminal and a site for palmitoylation in the C terminal. Encephalopsin displays both of these motifs. Although all opsins possess these motifs, they are not diagnostic for opsins, because other G-protein-linked receptors also possess such sites.
Comparison of the encephalopsin sequence with that of many other opsins and related proteins indicates that encephalopsin is a member of the vertebrate opsin family (Fig. 2). Resemblance of encephalopsin to nonopsin G-protein-coupled receptors and to retinochrome is substantially less than is resemblance to both vertebrate and invertebrate opsins. A unique two amino acid gap corresponding to residues 200 and 201 in bovine rhodopsin that occurs only in extraretinal vertebrate opsins (Okano et al., 1994; Max et al., 1995; Blackshaw and Snyder, 1997b; Soni and Foster, 1997; Sun et al., 1997; Provencio et al., 1998) and in invertebrate opsins is present in encephalopsin. Mouse and human encephalopsins are highly homologous to one another, showing 85.5% amino acid identity and 92% amino acid similarity over the full-length of the protein.
Fig. 2.
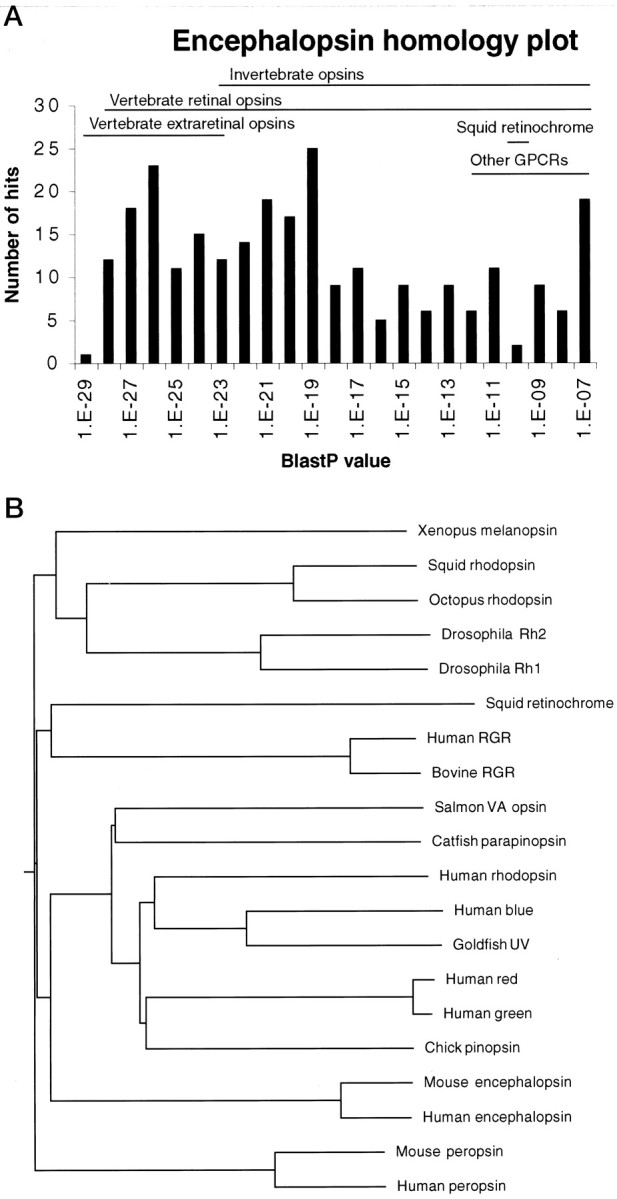
A, P value distribution of matches to mouse encephalopsin matches determined by a BLASTP search using BLAST 2.0 against the December 1998 GenBank release. Redundant sequences were not eliminated for this analysis. Other GPCRs represent nonopsin G-protein-coupled receptors. B, Dendrogram of encephalopsin phylogeny. Dendrograms are determined by CLUSTALW analysis. The gap penalty is set at 2, and the gap extension penalty is set at 0.01. Encephalopsin groups with vertebrate retinal and pineal opsins.
Analysis by a dendrogram distinguishes two large families, vertebrate and invertebrate opsins (Fig. 2B). Mouse and human encephalopsins clearly belong to the vertebrate family.
Attempts to reconstitute biochemically (Merbs and Nathans, 1992) encephalopsin in transfected 293 cells and efforts to demonstrate light-activated calcium influx in mammalian cells cotransfected with Gα16 (Offermanns and Simon, 1995; Krautwurst et al., 1998) and encephalopsin have not resulted in reproducible evidence of light absorption (data not shown).
Encephalopsin is enriched in brain and testes and expressed in putative light-sensitive areas of the diencephalon
Northern blot analysis reveals expression of encephalopsin only in cortex, cerebellum, and testes (Fig. 3). With longer exposures, we observe expression in whole brain, along with some faint expression in heart, liver, and kidney (data not shown). Even with very prolonged exposure, we observe no expression of encephalopsin in the retina, pineal gland, or skin. RT-PCR analysis also fails to reveal any encephalopsin in these three areas (data not shown).
Fig. 3.
Northern blot analysis of mouse encephalopsin expression. A single band of 1.9 kb is detected.
In situ hybridization shows a striking selectivity of encephalopsin expression in the adult mouse brain (Fig.4). Very high densities are evident in lateral regions of the thalamus, the cerebellum, and portions of the frontal cortex (Fig. 4A). Within the cerebral cortex, expression is predominant in frontal, perirhinal, cingulate, and insular cortex (Fig. 4B). Only negligible expression is evident in the parietal, temporal, and occipital cortices. Within the thalamus, encephalopsin expression is highest in the VPL, the posterior thalamus, the dorsomedial thalamus, and the lateral and medial geniculate nuclei, with much lower expression in mediodorsal and other more medially located nuclei of the thalamus. Within the cerebellum, expression is exclusively localized to Purkinje cells. Very high expression is also evident in the anteromedial preoptic area and subfornical organ.
Fig. 4.
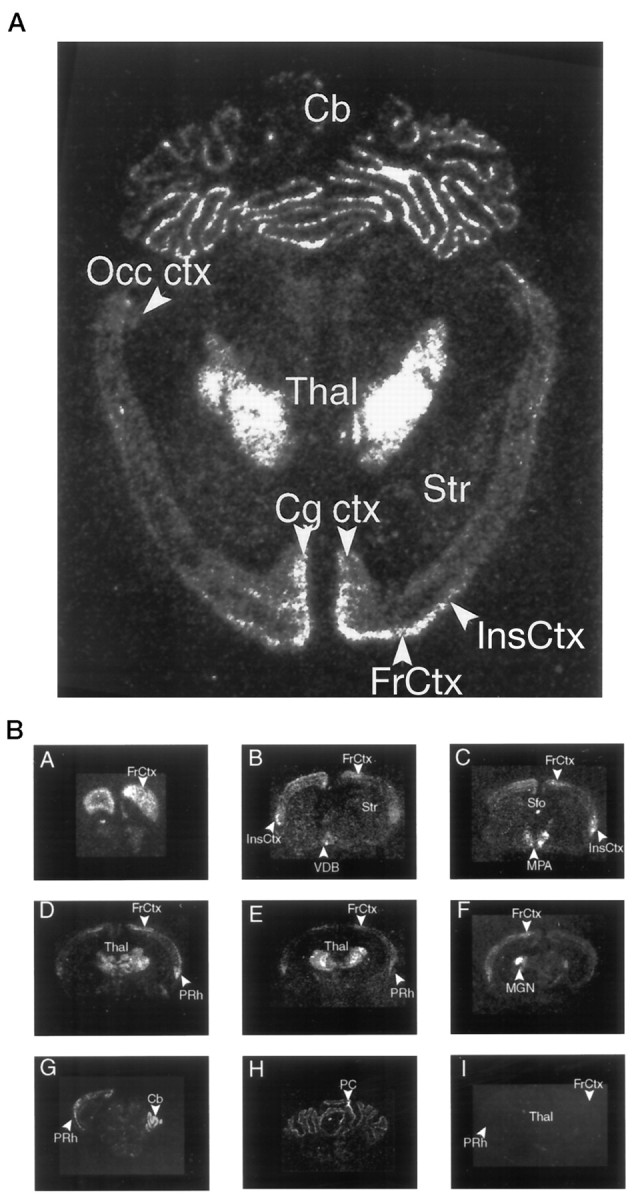
A, Expression of encephalopsin in a horizontal section of adult mouse brain. Radioactive in situ hybridization is used. B, Expression of encephalopsin in a rostrocaudal series of coronal sections of adult mouse brain. Radioactive in situ hybridization is used.Cb, Cerebellum; Cg ctx, cingulate cortex;FrCtx, frontal cortex; InsCtx, insular cortex; MPA, medial preoptic area; Occ ctx, occipital cortex; PC, Purkinje cell of cerebellum; PRh, perirhinal cortex; Sfo, subfornical organ; Str, striatum; Thal, thalamus; VDB, ventral diagonal band.
The most extensive evidence of light detection in the brain has been reported in selected nuclei in the diencephalon, specifically the paraventricular nuclei and the anterior medial preoptic area (Foster et al., 1994; Vigh and Vigh-Teichmann, 1998). Within the diencephalon, it is notable that the paraventricular nuclei and the anterior medial preoptic areas selectively express high levels of encephalopsin (Fig.5).
Fig. 5.
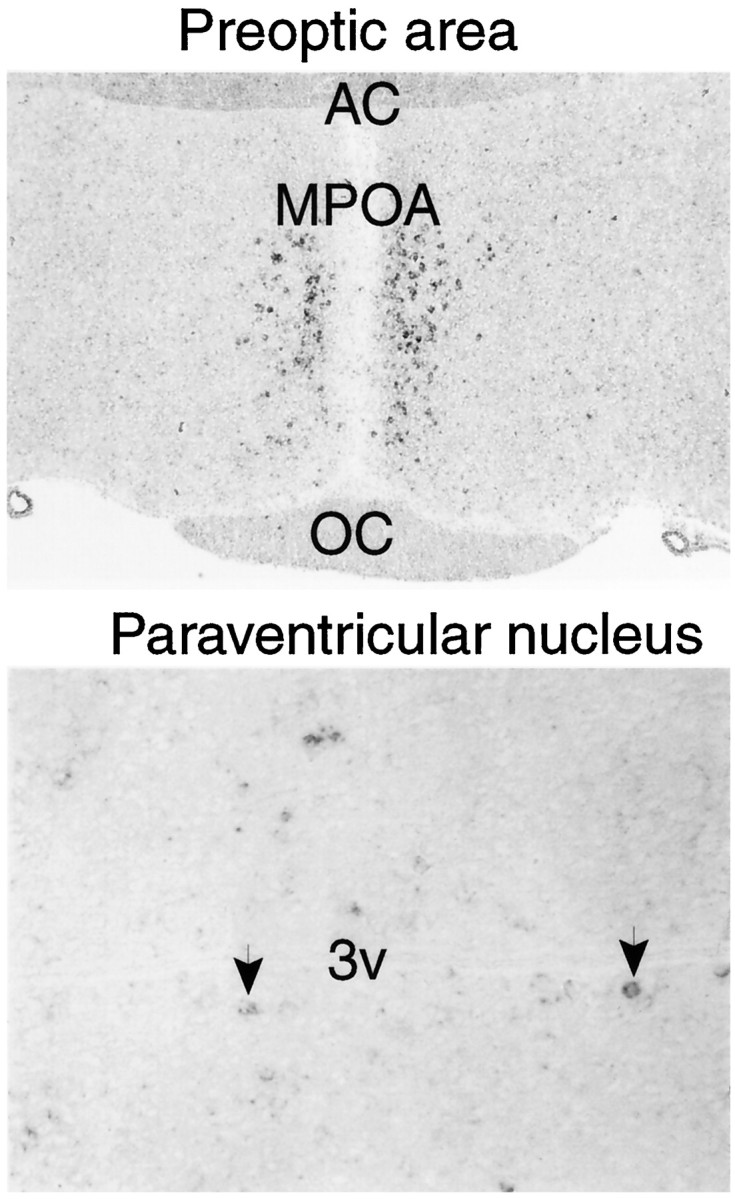
Digoxygenin in situ hybridization of encephalopsin expression in diencephalic structures.Top, The preoptic nucleus is taken at 50×.Bottom, The paraventricular nucleus is at 200×.Arrows indicate selected encephalopsin-positive cells in the paraventricular nucleus of the hypothalamus. AC, Anterior commissure; MPOA, medial preoptic area;OC, optic chiasm; 3v, third ventricle.
Encephalopsin displays rostrocaudal gradients of expression
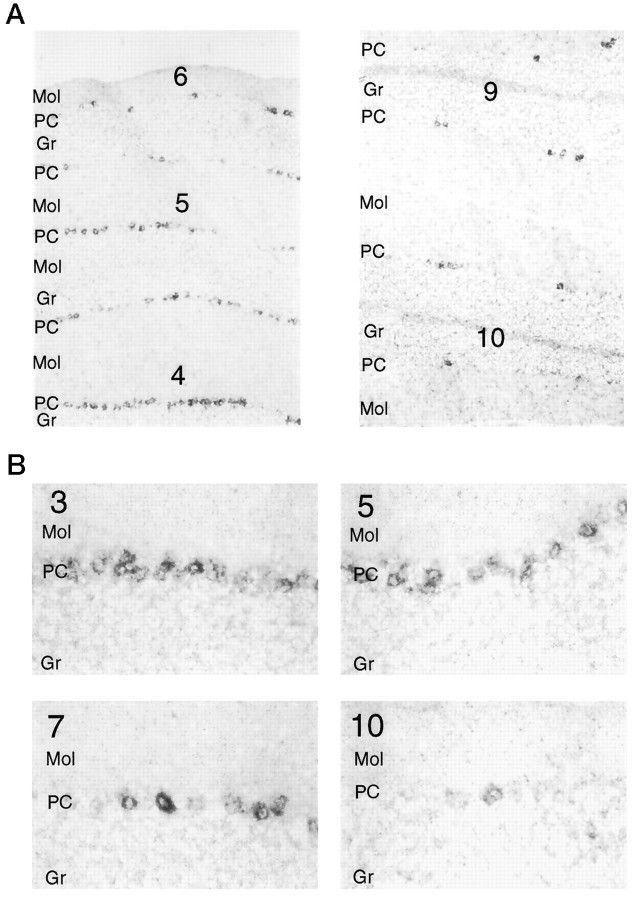
In several areas of the brain we observe striking rostrocaudal gradients of encephalopsin expression (Fig.6A). Within the cer- ebellum, the more anteriorly located lobes, such as 4, 5, and 6, are more enriched in encephalopsin than are the posteriorly located lobes, such as 8–10. Both the numbers of encephalopsin-positive cells and the intensity with which they express encephalopsin vary along the rostrocaudal axis (Fig. 6B). In cerebral cortex, encephalopsin expression is most prominent in layer 4, with somewhat less expression in 5 and very little expression in 6 (Fig. 7A). This lower expression is greatest in the frontal, cingulate, and insular cortices and much less in the periglomerular cortex. Expression is greater in rostral than caudal areas of these portions of the cortex. In the corpus striatum, encephalopsin is expressed in the population of small neurons whose density is greater in rostral than caudal zones (Fig. 7B).
Fig. 6.
A, Encephalopsin shows a rostrocaudal gradient of expression in the cerebellum. Digoxygeninin situ hybridization is used. Numbersindicate the lobe of the vermis and are numbered in rostrocaudal order. Pictures are taken at 50×. B, Both the number of encephalopsin-expressing cells and the intensity of encephalopsin expression show a rostrocaudal gradient in the cerebellum. Pictures are taken at 400×. Gr, Granule cell layer;Mol, molecular layer; PC, Purkinje cell layer.
Fig. 7.
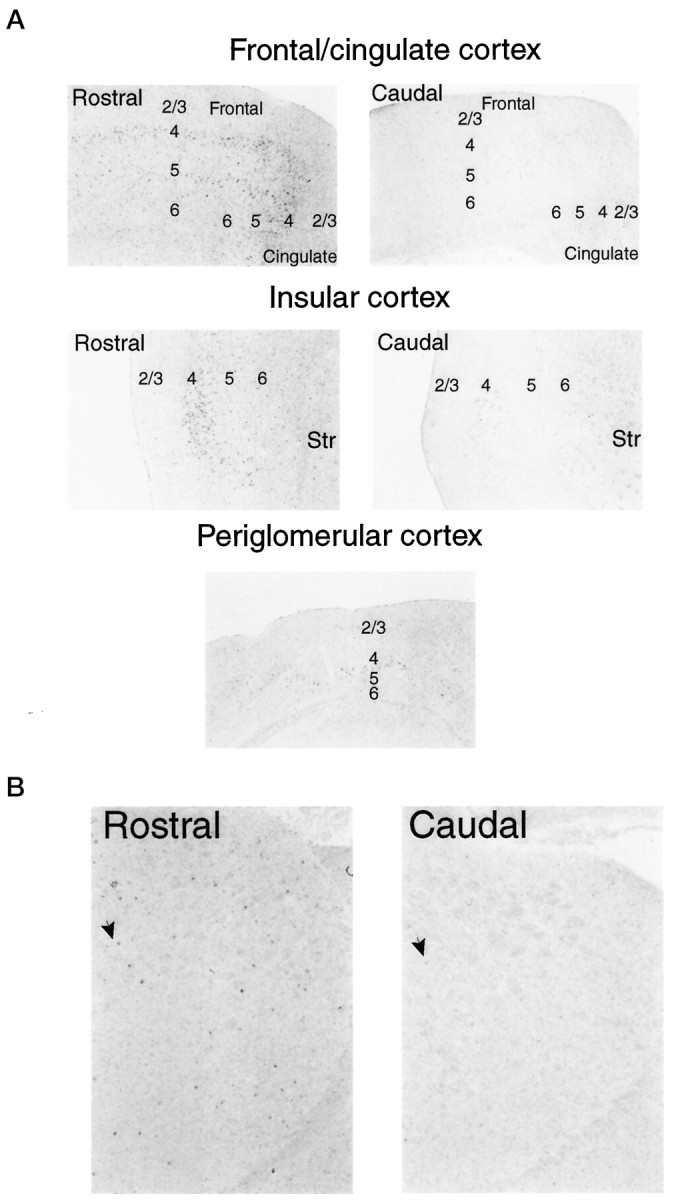
A, Encephalopsin expression shows a rostrocaudal gradient within the cerebral cortex. Cortical layers are indicated by numbers. Digoxygenin in situhybridization is used. All pictures are taken at 50×.B, Encephalopsin expression shows a rostrocaudal expression gradient in the striatum. Arrows indicate selected encephalopsin-positive cells. Pictures are taken at 50×.Str, Striatum.
Cerebellar encephalopsin displays a striped pattern of expression
At low magnification in the cerebellum, encephalopsin expression occurs in interrupted lines in both a horizontal and vertical dimension and in both the vermis and hemisphere (Fig.8A). Expression is evident only in Purkinje cells. Analysis at higher power reveals that this expression involves radially oriented stripes that are evident in both vertical and horizontal dimensions (Fig.8B,C). Examination of consecutive sections, focusing on a single point indicated by anarrow (Fig. 8B–D), reveals the appear- ance and disappearance of negative and positive stripes. In the hemispheres the stripes are wider than in the vermis (Fig.8D).
Fig. 8.
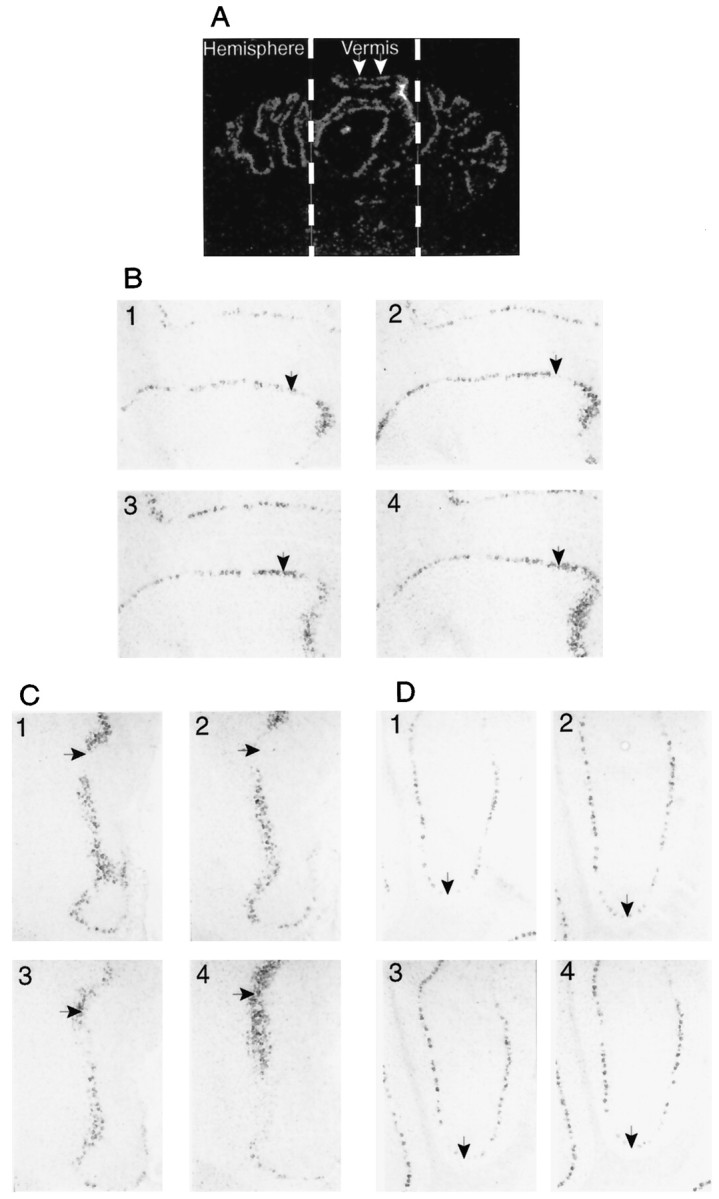
A, Encephalopsin is expressed in a striped pattern in the cerebellum. Radioactive in situhybridization is used on coronal sections. The vermis and hemispheres are indicated. White arrows indicate two consecutive stripes. B, Horizontal stripes of encephalopsin expression in the vermis are shown. Digoxygenin in situhybridization is used here. Consecutive sections are shown and numbered accordingly. All pictures are taken at 50×. C, Vertical stripes of encephalopsin expression are seen in the vermis. The pictures shown are from the same adjacent sections used inB. All pictures are taken at 50×. D, Stripes of encephalopsin expression in the cerebellar hemispheres are shown. Pictures are taken from the same series of consecutive sections used in B and C. All pictures are taken at 50×.
Encephalopsin expression is developmentally regulated
We fail to see any expression of encephalopsin in the embryonic cerebellum and cerebral cortex, and levels in the adult cerebellum and cortex are substantially higher than are those in the early postnatal period. Additionally, the rostrocaudal gradients and the cerebellar stripes are not clearly evident until 3 weeks of age (Fig.9A). At embryonic day 18.5 (E18.5), no encephalopsin expression is detected in the cerebellum despite substantial levels of encephalopsin in the adjacent dorsal pons. At postnatal day 4 (P4), Purkinje cells express encephalopsin, but no stripes or gradients are clearly evident. At P20, stripes can be seen, but these are substantially more notable in the adult cerebellum. This developmental pattern is particularly striking in light of a very different developmental pattern for most other forms of cerebellar stripes. Of the proteins expressed in a striped pattern in the cerebellum, most are prominent in embryonic and early postnatal life, and many are thought to play a role in the development of the cerebellum (Herrup and Kuemerle, 1997; Lin and Cepko, 1998; Oberdick et al., 1998). Encephalopsin is unusual in its exclusive expression as stripes in the adult but not in earlier stages.
Fig. 9.
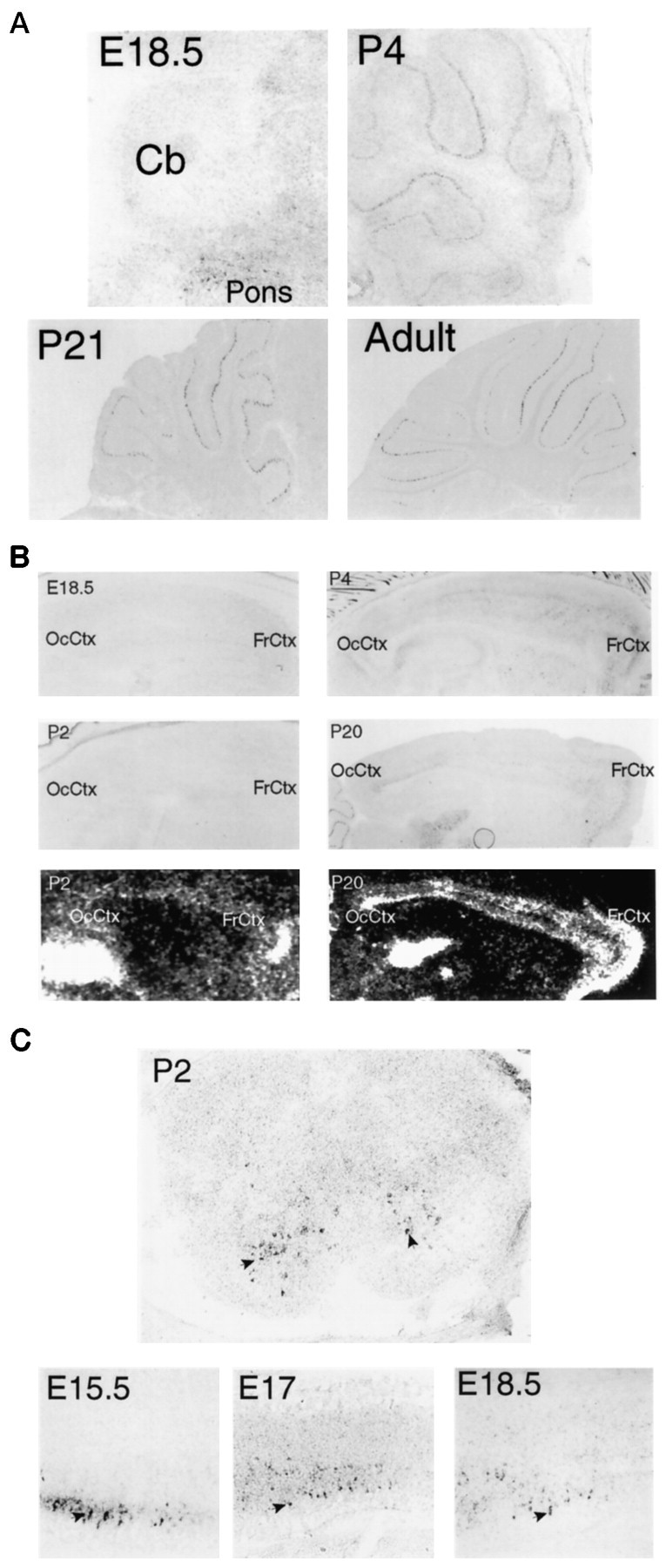
A, Developmental expression of encephalopsin in the cerebellum is shown. The adult pattern of encephalopsin expression is not seen until several weeks after birth. Sagittal sections are shown. E18.5 is taken at 100×, P4 is taken at 50×, and P21 and adult are taken at 25×. B, Developmental expression of encephalopsin expression in the cerebral cortex is shown. Encephalopsin expression is absent prenatally, relatively evenly distributed throughout the cortex in the early postnatal mouse, and showing rostrocaudal gradients and regional organization by P20. Digoxy genin in situ hybridization is shown for E18.5, P2, P4, and P20, whereas radioactive in situhybridization (bottom) is shown for P2 and P20. Digoxygenin in situ hybridization pictures are taken at 50× for E18.5, 25× for P2 and P4, and 12.5× for P20.C, Encephalopsin expression is prominent in developing spinal cord. Sagittal sections of embryonic time points are taken of the cervical cord at 100× for E15.5 and 50× for E17 and E18.5. The coronal section at P2 is taken of cervical spinal cord at 50×.Arrows indicate selected encephalopsin-positive interneurons of the ventral horn. The color reaction in the P2 section is allowed to proceed twice as long as that in the embryonic time points, so the observed abundance levels of encephalopsin are not directly comparable. Cb, Cerebellum;FrCtx, frontal cortex; OcCtx, occipital cortex.
In the cerebral cortex, the rostrocaudal gradient is absent at E18.5, P2, and P4 but becomes evident at P20 and is most prominent in the adult (Fig. 9B).
In contrast to the negligible expression of encephalopsin in embryonic life in most parts of the nervous system, the spinal cord displays substantial encephalopsin at early stages (Fig. 9C). The highest levels of encephalopsin occur in a subset of interneurons in the ventral cord of the spinal cord at E15.5. Expression decreases at E17 and E18. The spinal cord reaches functional maturity at birth. At P2, encephalopsin expression is clearly restricted to the area of interneurons in the ventral horn.
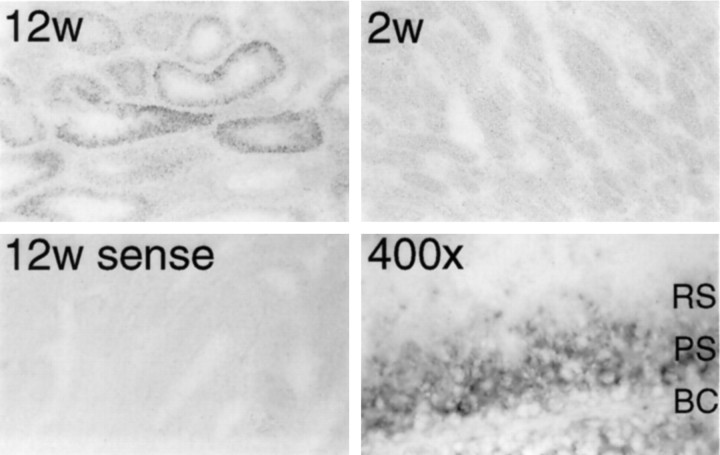
Encephalopsin is localized to pachytene spermatids in the testes
In situ hybridization reveals developmental changes in encephalopsin expression in the testes (Fig.10). No expression is evident at 2 weeks of age, whereas substantial expression occurs at 12 weeks when mature sperm are forming. Only a subset of seminiferous tubules expresses encephalopsin. At high power, encephalopsin expression is restricted to pachytene spermatids with negligible levels in basal cells or in the mature round spermatids. Pachytene spermatids are undergoing meiosis. During meiosis, marked upregulation takes place for many genes unrelated to the meiotic process, and many of these transcripts are not translated into protein (Eddy and O’Brien, 1998). Thus, it is unclear whether encephalopsin in these spermatids plays a functional role. The specificity of the in situhybridization process is indicated by the absence of staining with sense control probes.
Fig. 10.
Strong encephalopsin expression is observed in meiotic spermatids. Encephalopsin expression is seen in a subset of seminiferous tubules of the 12-week-old (12w) but not the 2 week-old (2w) mouse, indicating expression in sperm. High-power pictures indicate that encephalopsin expression is restricted to pachytene spermatids. All pictures are at 100×, except for the high-power shot that is at 400×. BC, Basal cells; PS, pachytene spermatids; RS, round spermatids.
DISCUSSION
The major finding of our study is the identification and characterization of what appears to be the first opsin expressed specifically in the mammalian brain. Until now mammalian opsins have been described as specifically expressed only in the retina and the pineal. In a few instances, expression of certain retinal and pineal opsins has been detected in selected regions of lower vertebrate brains.
Because the major function of opsins involves light detection, we must consider the possibility that encephalopsin participates in such a process. There is a substantial literature establishing the presence of photoreceptors in the diencephalon of several lower vertebrates (Menaker et al., 1970; Hartwig and Oksche, 1982). Although light entrainment of circadian rhythms in adult mammals requires the presence of intact eyes (Menaker et al., 1997; Roenneberg and Foster, 1997), this does not rule out the presence of extraocular photoreceptors that may play a modulatory role in circadian entrainment (Foster, 1998). Abundant evidence establishes that light can penetrate through the skull into deep layers of the mammalian brain including the diencephalon (van Brundt et al., 1964; Wurtman, 1975; Hartwig and van Veen, 1979). These studies have used both rodents (Viggiani et al., 1970) and humans (Muller and Wilson, 1986). Interestingly, direct effects of light on functions of the cerebral cortex have been reported (Wade et al., 1988), with light application to cerebral cortical slices regulating neurotransmitter release.
How do the properties of encephalopsin fit with a potential role in light perception? The molecular architecture of encephalopsin indicates that it possesses all the appropriate residues for phototransduction, most notably the lysine at position 297 and the counterion at residue 115. In preliminary experiments, we attempted to reconstitute encephalopsin biochemically and to monitor light absorption. However, we have so far been unable to obtain reproducible evidence of light absorption. Similar attempts to reconstitute recombinant extraretinal opsins or novel opsins expressed in the mammalian retinal pigmented epithelium have typically been unsuccessful, except for only pinopsin (Okano et al., 1994) and VA opsin (Soni et al., 1998), both of which are considerably more homologous to retinal opsins than is encephalopsin.
Some of the localizations of encephalopsin in the brain fit with the literature on brain phototransduction. Thus, within the diencephalon, encephalopsin is enriched in the paraventricular nucleus and the medial preoptic nucleus, both of which are thought to play a role in encephalic photoreception (Vigh et al., 1983; Foster et al., 1994; Vigh and Vigh-Teichmann, 1998). Other localizations, such as the Purkinje cells of the cerebellum and various neurons in the cerebral cortex and elsewhere, have no known relationship to circadian photoreception. Interestingly, the cerebellum and cerebral cortex are substantially more superficial structures than is the diencephalon and hence would have greater access to light. Finally, even in the absence of light activation, extraretinal opsins may form some essential component of the circadian clock, as has been suggested by the discovery of functional opsins in eyeless cave-dwelling fish (Tabata, 1982) and crustaceans (Crandall and Hillis, 1997).
Conceivably, encephalopsin is a G-protein-coupled retinoid receptor. The known retinoid receptors do not resemble opsin in structure and are instead all transcription factors of the nuclear hormone receptor superfamily. They are notably involved in processes related to development, such as early patterning and cell fate (Durston et al., 1998; Maden, 1998). Rostrocaudal variations in protein expression and stripes in the cerebellum are often associated with these types of developmental events. However, in most cases the proteins displaying these properties are highly expressed early in development (Herrup and Kuemerle, 1997; Oberdick et al., 1998). By contrast, encephalopsin expression is much higher and more elaborately patterned in the adult. Accordingly, it would seem more likely that the rostrocaudal gradients and cerebellar stripes participate in some coding mechanism. In the mammalian retina, gradients of rhodopsin and cone protein expression are seen and are thought to play a role in visual processing (Wang et al., 1992; Cepko, 1996). Similar gradients of opsin expression also occur in photosensitive pineal organs (Blackshaw and Snyder, 1997b). Gradients of encephalopsin in the mammalian brain could be serving an analogous function.
Footnotes
This work was supported by United States Public Health Service Grant DA-00268 and Research Scientist Award DA-00074 to S.H.S. We thank H. Sun and D. Krautwurst for assistance with the biochemical reconstitution of encephalopsin, J. Nathans and K.-W. Yau for comments on this manuscript, and J. Pevsner, D. Linden, and C. Riley for helpful discussions.
Correspondence should be addressed to Dr. Solomon H. Snyder, The Johns Hopkins University, School of Medicine, Department of Neuroscience, 725 North Wolfe Street, Baltimore, MD 21205.
Dr. Blackshaw’s present address: Department of Genetics, Harvard Medical School, 200 Longwood Avenue, Boston, MA 02115.
REFERENCES
- 1.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 2.Blackshaw S, Snyder SH. Developmental expression pattern of phototransduction components in mammalian pineal implies a light-sensing function. J Neurosci. 1997a;17:8074–8082. doi: 10.1523/JNEUROSCI.17-21-08074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackshaw S, Snyder SH. Parapinopsin, a novel catfish opsin localized to the parapineal organ, defines a new gene family. J Neurosci. 1997b;17:8083–8092. doi: 10.1523/JNEUROSCI.17-21-08083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell SS, Murphy PJ. Extraocular circadian phototransduction in humans. Science. 1998;279:396–399. doi: 10.1126/science.279.5349.396. [DOI] [PubMed] [Google Scholar]
- 5.Cepko CL. The patterning and onset of opsin expression in vertebrate retinae. Curr Opin Neurobiol. 1996;6:542–546. doi: 10.1016/s0959-4388(96)80062-6. [DOI] [PubMed] [Google Scholar]
- 6.Crandall KA, Hillis DM. Rhodopsin evolution in the dark. Nature. 1997;387:667–668. doi: 10.1038/42628. [DOI] [PubMed] [Google Scholar]
- 7.Durston AJ, van der Wees J, Pijnappel WW, Godsave SF. Retinoids and related signals in early development of the vertebrate central nervous system. Curr Top Dev Biol. 1998;40:111–175. doi: 10.1016/s0070-2153(08)60366-x. [DOI] [PubMed] [Google Scholar]
- 8.Eddy EM, O’Brien DA. Gene expression during mammalian meiosis. Curr Top Dev Biol. 1998;37:141–200. [PubMed] [Google Scholar]
- 9.Foster RG. Shedding light on the biological clock. Neuron. 1998;20:829–832. doi: 10.1016/s0896-6273(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 10.Foster RG, Grace MS, Provencio I, Degrip WJ, Garcia-Fernandez JM. Identification of vertebrate deep brain photoreceptors. Neurosci Biobehav Rev. 1994;18:541–546. doi: 10.1016/0149-7634(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 11.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green CB. How cells tell time. Trends Cell Biol. 1998;8:224–230. doi: 10.1016/s0962-8924(98)01269-0. [DOI] [PubMed] [Google Scholar]
- 13.Hara-Nishimura I, Matsumoto T, Mori H, Nishimura M, Hara R, Hara T. Cloning and nucleotide sequence of cDNA for retinochrome, retinal photoisomerase from the squid retina. FEBS Lett. 1990;271:106–110. doi: 10.1016/0014-5793(90)80383-t. [DOI] [PubMed] [Google Scholar]
- 14.Hartwig HG, Oksche A. Neurobiological aspects of extraretinal photoreceptive systems: structure and function. Experientia. 1982;38:991–996. doi: 10.1007/BF01955341. [DOI] [PubMed] [Google Scholar]
- 15.Hartwig H-G, van Veen T. Spectral characteristics of visible radiation penetrating into the brain and stimulating extraretinal photoreceptors. J Comp Physiol [A] 1979;130:277–282. [Google Scholar]
- 16.Herrup K, Kuemerle B. The compartmentalization of the cerebellum. Annu Rev Neurosci. 1997;20:61–90. doi: 10.1146/annurev.neuro.20.1.61. [DOI] [PubMed] [Google Scholar]
- 17.Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 18.Lin JC, Cepko CL. Granule cell raphes and parasagittal domains of Purkinje cells: complementary patterns in the developing chick cerebellum. J Neurosci. 1998;18:9342–9353. doi: 10.1523/JNEUROSCI.18-22-09342.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin SW, Kochendoerfer GG, Carroll KS, Wang D, Mathies RA, Sakmar TP. Mechanisms of spectral tuning in blue cone visual pigments. Visible and raman spectroscopy of blue-shifted rhodopsin mutants. J Biol Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- 20.Maden M. Vertebrate development: a nervous vitamin. Curr Biol. 1998;8:R846–R849. doi: 10.1016/s0960-9822(07)00528-3. [DOI] [PubMed] [Google Scholar]
- 21.Max M, McKinnon PJ, Seidenman KJ, Barrett RK, Applebury ML, Takahashi JS, Margolskee RF. Pineal opsin: a nonvisual opsin expressed in chick pineal. Science. 1995;267:1502–1506. doi: 10.1126/science.7878470. [DOI] [PubMed] [Google Scholar]
- 22.Menaker M, Roberts R, Elliott J, Underwood H. Extraretinal light perception in the sparrow. 3. The eyes do not participate in photoperiodic photoreception. Proc Natl Acad Sci USA. 1970;67:320–325. doi: 10.1073/pnas.67.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menaker M, Moreira LF, Tosini G. Evolution of circadian organization in vertebrates. Braz J Med Biol Res. 1997;30:305–313. doi: 10.1590/s0100-879x1997000300003. [DOI] [PubMed] [Google Scholar]
- 24.Merbs SL, Nathans J. Absorption spectra of human cone pigments. Nature. 1992;356:433–435. doi: 10.1038/356433a0. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto Y, Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller PJ, Wilson BC. An update on the penetration depth of 630 nm light in normal and malignant human brain tissue in vivo. Phys Med Biol. 1986;31:1295–1297. doi: 10.1088/0031-9155/31/11/012. [DOI] [PubMed] [Google Scholar]
- 27.Nathans J. Determinants of visual pigment absorbance: identification of the retinylidene Schiff’s base counterion in bovine rhodopsin. Biochemistry. 1990;29:9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- 28.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 29.Neitz M, Neitz J, Jacobs GH. Spectral tuning of pigments underlying red-green color vision. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- 30.Oberdick J, Baader SL, Schilling K. From zebra stripes to postal zones: deciphering patterns of gene expression in the cerebellum. Trends Neurosci. 1998;21:383–390. doi: 10.1016/s0166-2236(98)01325-3. [DOI] [PubMed] [Google Scholar]
- 31.Offermanns S, Simon MI. G alpha 15 and G alpha 16 couple a wide variety of receptors to phospholipase C. J Biol Chem. 1995;270:15175–15180. doi: 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- 32.Okano T, Yoshizawa T, Fukada Y. Pinopsin is a chicken pineal photoreceptive molecule. Nature. 1994;372:94–97. doi: 10.1038/372094a0. [DOI] [PubMed] [Google Scholar]
- 33.Palczewski K. Is vertebrate phototransduction solved? New insights into the molecular mechanism of phototransduction. Invest Ophthalmol Vis Sci. 1994;35:3577–3581. [PubMed] [Google Scholar]
- 34.Pang K, Knecht D. Partial inverse PCR: a technique for cloning flanking sequences. Biotechniques. 1997;22:1046–1048. doi: 10.2144/97226bm07. [DOI] [PubMed] [Google Scholar]
- 35.Provencio I, Foster RG. Circadian rhythms in mice can be regulated by photoreceptors with cone-like characteristics. Brain Res. 1995;694:183–190. doi: 10.1016/0006-8993(95)00694-l. [DOI] [PubMed] [Google Scholar]
- 36.Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: an opsin in melanophores, brain, and eye. Proc Natl Acad Sci USA. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roenneberg T, Foster RG. Twilight times: light and the circadian system. Photochem Photobiol. 1997;66:549–561. doi: 10.1111/j.1751-1097.1997.tb03188.x. [DOI] [PubMed] [Google Scholar]
- 38.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schertler GF. Structure of rhodopsin. Eye. 1998;12:504–510. doi: 10.1038/eye.1998.138. [DOI] [PubMed] [Google Scholar]
- 40.Shen D, Jiang M, Hao W, Tao L, Salazar M, Fong HK. A human opsin-related gene that encodes a retinaldehyde-binding protein. Biochemistry. 1994;33:13117–13125. doi: 10.1021/bi00248a022. [DOI] [PubMed] [Google Scholar]
- 41.Soni BG, Foster RG. A novel and ancient vertebrate opsin. FEBS Lett. 1997;406:279–283. doi: 10.1016/s0014-5793(97)00287-1. [DOI] [PubMed] [Google Scholar]
- 42.Soni BG, Philp AR, Foster RG, Knox BE. Novel retinal photoreceptors. Nature. 1998;394:27–28. doi: 10.1038/27794. [DOI] [PubMed] [Google Scholar]
- 43.Sun H, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc Natl Acad Sci USA. 1997;94:9893–9898. doi: 10.1073/pnas.94.18.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabata M. Persistence of pineal photosensory function in blind cave fish, Astyanax mexicanus. Comp Biochem Physiol. 1982;73A:125–127. [Google Scholar]
- 45.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 46.van Brundt EE, Shepherd MD, Wale JR, Ganong WF, Clegg M-T. Penetration of light into the brain of mammals. Ann NY Acad Sci. 1964;117:217–224. doi: 10.1111/j.1749-6632.1964.tb48177.x. [DOI] [PubMed] [Google Scholar]
- 47.Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 48.Viggiani E, Ciesla M, Russo OL. The shielding power of the rat skull to visible light. Experientia. 1970;26:850–851. doi: 10.1007/BF02114217. [DOI] [PubMed] [Google Scholar]
- 49.Vigh B, Vigh-Teichmann I. Actual problems of the cerebrospinal fluid-contacting neurons. Microsc Res Tech. 1998;41:57–83. doi: 10.1002/(SICI)1097-0029(19980401)41:1<57::AID-JEMT6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 50.Vigh B, Vigh-Teichmann I, Rohlich P, Oksche A. Cerebrospinal fluid-contacting neurons, sensory pinealocytes and Landolt’s clubs of the retina as revealed by means of an electron-microscopic immunoreaction against opsin. Cell Tissue Res. 1983;233:539–548. doi: 10.1007/BF00212223. [DOI] [PubMed] [Google Scholar]
- 51.Wada Y, Okano T, Adachi A, Ebihara S, Fukada Y. Identification of rhodopsin in the pigeon deep brain. FEBS Lett. 1998;424:53–56. doi: 10.1016/s0014-5793(98)00138-0. [DOI] [PubMed] [Google Scholar]
- 52.Wade PD, Taylor J, Siekevitz P. Mammalian cerebral cortical tissue responds to low-intensity visible light. Proc Natl Acad Sci USA. 1988;85:9322–9326. doi: 10.1073/pnas.85.23.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, Bennett J, Gearhart J, Nathans J. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 54.Wurtman RJ. The effects of light on the human body. Sci Am. 1975;233:69–77. [PubMed] [Google Scholar]
- 55.Yokoyama S. Molecular evolution of retinal and nonretinal opsins. Genes Cells. 1996;1:787–794. doi: 10.1046/j.1365-2443.1996.d01-273.x. [DOI] [PubMed] [Google Scholar]
- 56.Yoshikawa T, Okano T, Oishi T, Fukada Y. A deep brain photoreceptive molecule in the toad hypothalamus. FEBS Lett. 1998;424:69–72. doi: 10.1016/s0014-5793(98)00139-2. [DOI] [PubMed] [Google Scholar]
- 57.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]