Fig. 3.
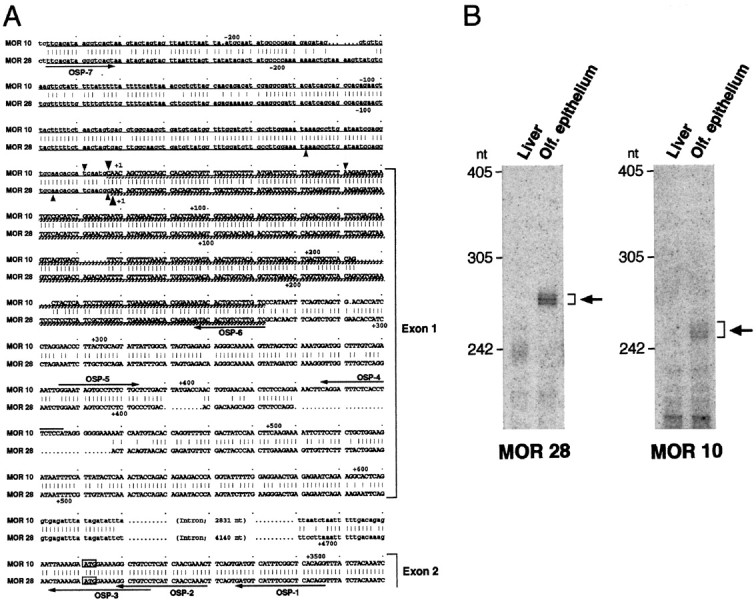
The 5′-noncoding sequences of the MOR10 and MOR28 transcripts. A, Nucleotide sequence comparison of the 5′-noncoding regions of the MOR10 and MOR28 genes. Identical nucleotides are connected by vertical bars, and both noncoding and coding sequences in exon 1 and exon 2 are depicted byuppercase letters. Upstream genomic sequences as well as intron sequences are noted by lowercase letters. The ATG initiation codons are boxed. Underlined sequences show regions used for antisense RNA probes in the RNase protection assay, and cross-hatched underlining sequences indicate protected regions. The major transcription start sites, identified by the 5′-RACE analysis and by the RNase protection assay, are marked bysmall and large arrowheads, respectively. Primers for the 5′-RACE and RT-PCR analyses are indicated byarrows along the sequences. B, The RNase protection assay. Antisense RNA probes labeled with 32P were hybridized to poly(A)+ RNAs from the mouse liver or olfactory epithelium. After hybridization, samples were digested with RNase and separated on a denaturing polyacrylamide gel (6%). In olfactory poly(A)+ RNA, three protected fragments of 247, 252, and 258 nt were detected for the MOR10 probe, whereas those of 269, 271, and 276 nt were detected for the MOR28 probe. No protected products were detected in liver mRNA.
