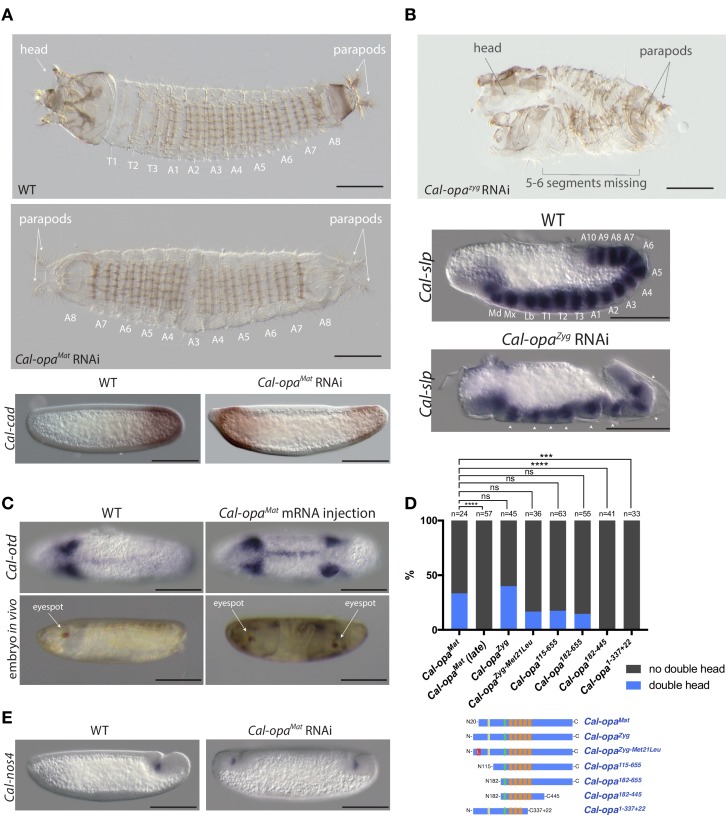
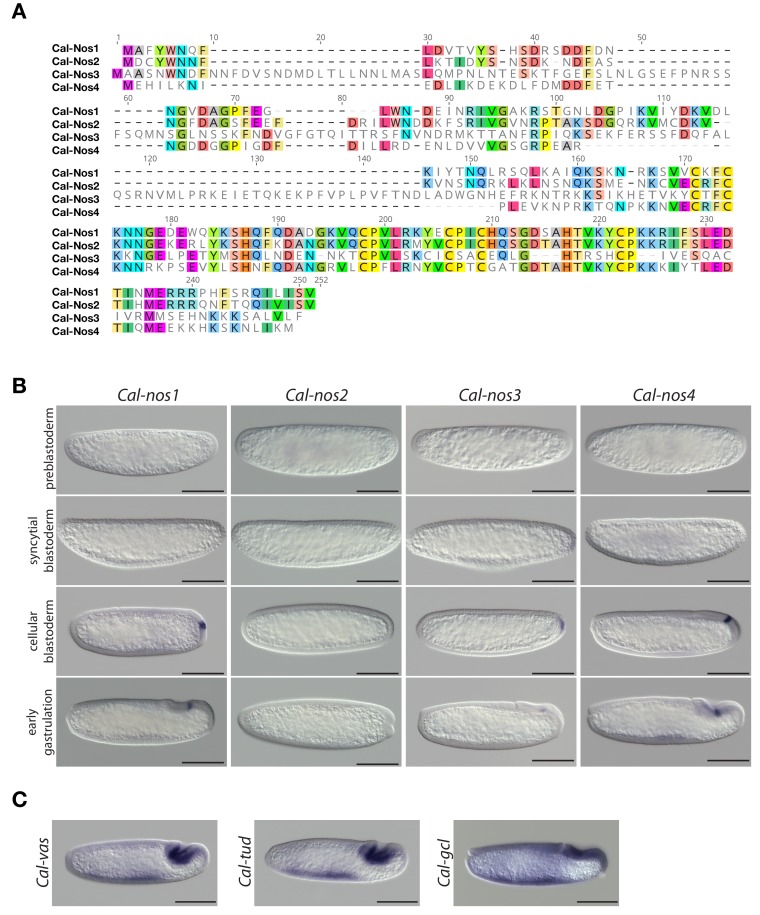
Figure 2. Function of alternative Cal-opa transcripts in early Clogmia embryo.
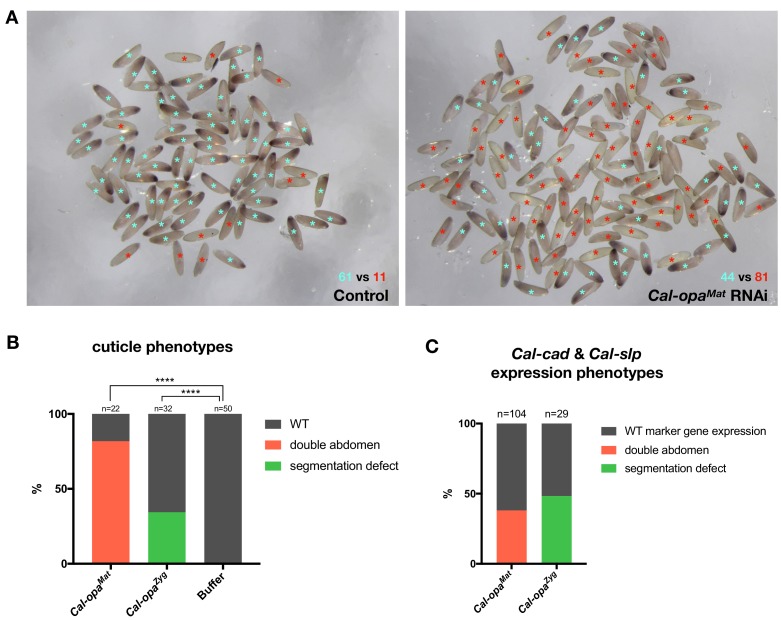
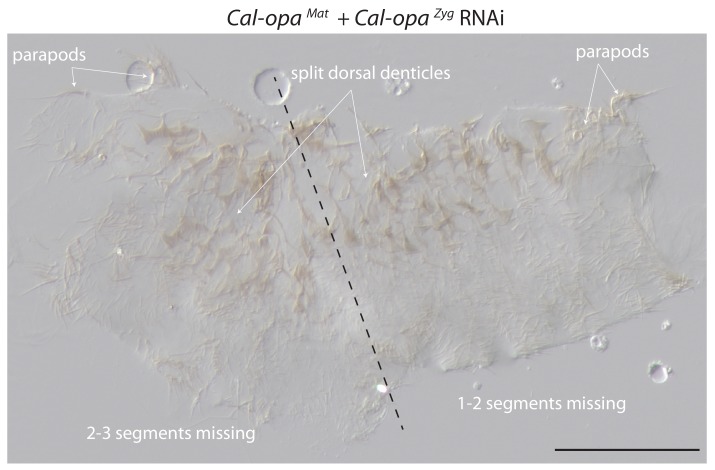
(A) 1 st instar larval cuticle of wild type (top) and following Cal-opaMat RNAi (middle). RNA in situ hybridization of Cal-cad in a wild-type preblastoderm embryo (bottom left) and stage-matched Cal-opaMat RNAi embryos (bottom right). Anterior is left and dorsal up. T: thoracic segment; A: abdominal segment. Segment numbers in Cal-opaMat RNAi larval cuticle were assigned based on the assumption of polarity reversal. Scale bar: 100μm. (B) 1 st instar larval cuticle phenotype following Cal-opaZyg RNAi (top) and RNA in situ hybridization of Cal-slp in extending wild-type germband (middle) and stage-matched Cal-opaZyg RNAi embryo (bottom). Anterior is left and dorsal up. Md: mandibular segment; Mx: maxillary segment; Lb: labial segment; T: thoracic segment; A: abdominal segment. Scale bar: 100μm. (C) RNA in situ hybridizations of Cal-otd in wild-type gastrula (top left) and stage-matched embryo following posterior Cal-opaMat mRNA injection (top right) are shown in ventral view. A live wild-type embryo (bottom left) and a stage-matched embryo following posterior Cal-opaMat mRNA injection (bottom right) in lateral view. Anterior is left. Scale bar: 100μm. (D) Posterior injection of Cal-opa mRNA and mutated variants. Complete, symmetrical duplication of the bilateral Cal-otd expression domain in gastrulating embryos was counted as double head (blue, see Figure 2C). All other phenotypes, including incomplete duplications and wild type, were conservatively counted as ‘no double head’ (black). Sketches of predicted Cal-Opa proteins are shown with ZIC/Opa conserved motif (ZOC) in yellow, the ZIC family protein N-terminal conserved domain (ZFNC) in green, and zinc finger domains in orange. The Met21Leu mutation in Cal-OpaZyg-Met21Leu is marked in red. Cal-opaMat (late): Cal-opaMat was injected during the syncytial blastoderm stage (4 hr). ns: p>0.05; ***: p<0.001; ****: p<0.0001, Fisher’s exact test. (E) RNA in situ hybridization of Cal-nos4 in a gastrulating embryo (left) and stage-matched Cal-opaMat RNAi embryos (right).