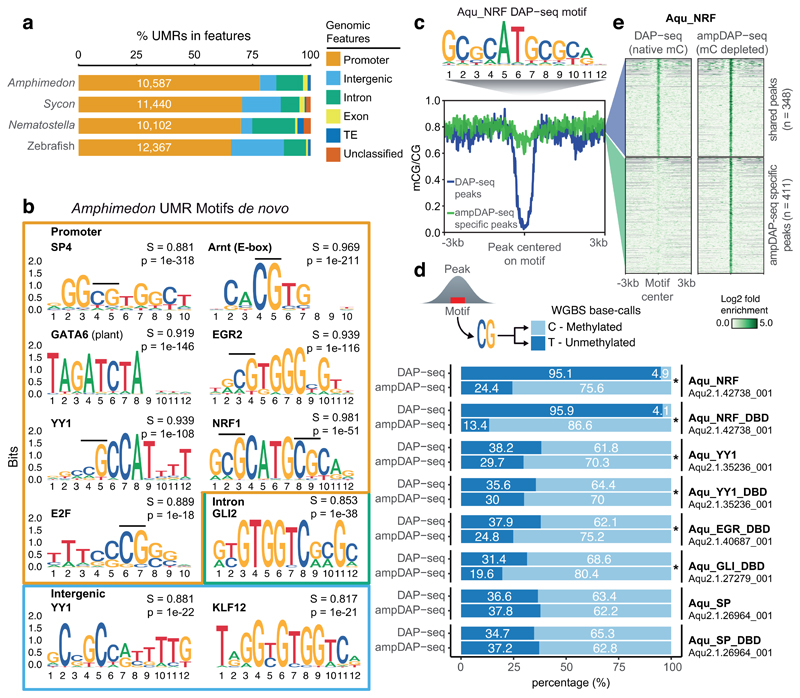
Figure 2. Methyl-sensitive transcription factors are enriched at unmethylated Amphimedon promoters.
(a) Unmethylated region (UMR) intersections with genomic features. Unclassified are regions found in scaffolds/contigs without any genes/transposable elements. (b) Sequence logos showing de novo motif enrichments for UMRs in promoters, introns and intergenic regions of Amphimedon. In bold is the best motif match, S is the match score to the known motif, p is the p-value enrichment (one-sided binomial test). (c) Motif enriched in Amphimedon NRF (Aqu_NRF) DAP-seq peaks. (d) Proportions of methylated calls versus unmethylated calls from Amphimedon whole genome bisulfite sequencing data at all DAP-seq peaks, and ampDAP-seq specific peaks. CpGs were obtained from the best motif within each peak. An asterisk shows significant enrichment for methylated calls in the ampDAP-seq specific peaks (one-sided Fisher exact test < 0.05). DBD (DNA Binding Domain). (e) Mean methylation profile at Amphimedon NRF DAP-seq peaks (blue) and ampDAP-seq specific peaks (green), and heatmap showing enrichment levels of NRF DAP-seq and ampDAP-seq at those peaks. DAP-seq and ampDAP-seq data shown as log2 fold enrichment of NRF against empty pIX-HALO plasmid.