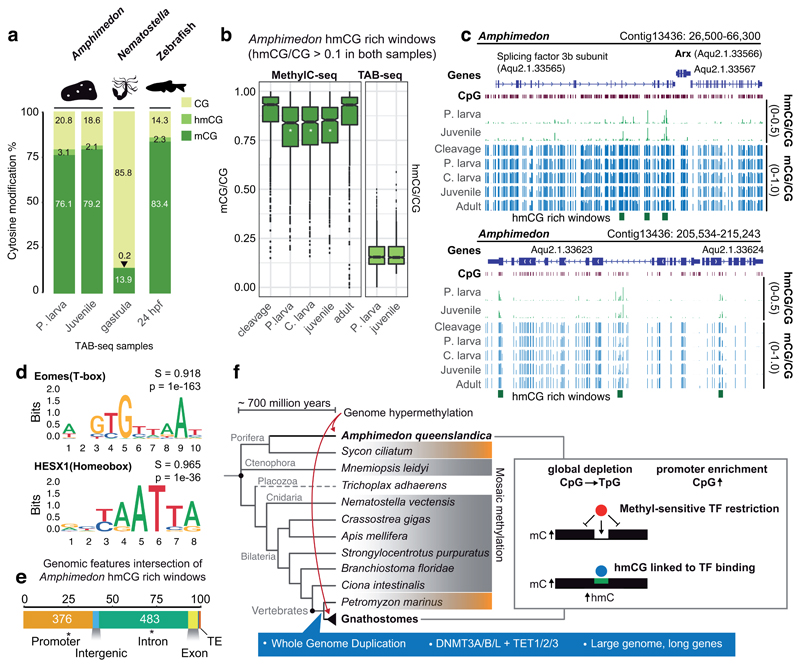
Figure 4. Genomic DNA hydroxymethylation is enriched at transcription factor binding sites in Amphimedon.
(a) Global mCG, hmCG and unmethylated CpG levels in Amphimedon, Nematostella and zebrafish assessed by MethylC-seq and TAB-seq corrected by non-conversion rate and protection rate. (b) Methylation and hydroxymethylation levels on hmCG rich windows (hmCG > 0.1 in both Precompetent larva and Juvenile stages). A white asterisk on the boxplot indicates FDR < 0.01 in a multiple one-sided wilcoxon enrichment test (Benjamini-Hochberg corrected) between the methylation level at hmCG rich windows against a set of random 150bp windows (n = 7,580) with equivalent coverage and CpG densities. Boxplot centre lines are medians, box limits are quartiles 1 (Q1) and 3 (Q3), whiskers are 1.5 × interquartile range (IQR) and points are outliers. (c) Genome browser representations of the Amphimedon Arx locus displaying distal hmC rich windows in the adjacent gene and transient demethylation in hmCG rich windows at mid-developmental samples (d) Sequence logos of de novo enriched motifs in Amphimedon hmCG rich windows. In bold is the best match to known motifs, S is the match score and p is the p-value enrichment (one-sided binomial test). (e) Intersection of genomic features with Amphimedon hmCG rich windows. Asterisk indicates two-sided Fisher’s exact test p < 0.05 enrichment against expected. (f) Cladogram representing phylogenetic relationships of animals. Species that have a sparse or “mosaic” DNA methylome are shaded in grey, those that have intermediate levels (~50%) are shaded in orange, and a dashed line indicates lack of methylation. The right-hand box summarizes the similarities observed between Amphimedon and vertebrate methylomes. In blue are listed some of the vertebrate characteristics that have been correlated to genome hypermethylation but are absent in Amphimedon.