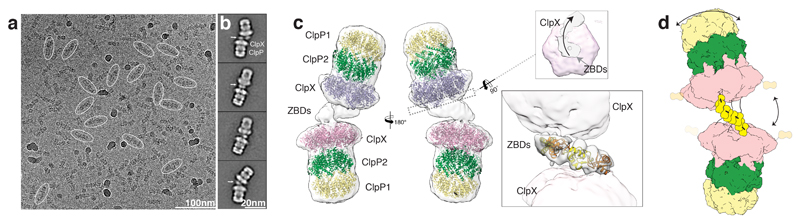
Figure 1. LmClpXP1/2 forms flexible dimers via the ZBDs.
a) Typical low-dose cryo-EM micrograph of the ClpXP1/2 dimer from L. monocytogenes. Some particles are highlighted with ovals. Scale bar, 100nm b) Typical reference-free 2D class averages. Arrows indicate additional densities corresponding to ZBDs at the interface between two ClpX hexamers. Scale bar, 20nm c) Ribbon Model of ClpP1 (yellow), ClpP2 (green) and ClpX (orange) superimposed with the cryo-EM density map of the ClpXP1/2 dimer (white and transparent). The upper inset shows the complex shown as slice at the position of the axial pore entry of the upper ClpXP1/2 complex. ClpX and ClpX-ZBD densities are colored magenta and gray transparent, respectively. The arrow indicates the spiral arrangement of the ZBD domains. The lower inset shows four copies of ZBD-dimers (PDB: 1OVX) placed into the cryo-EM density at the interface between the ClpX hexamers. The low resolution density did not allow automated rigid-body fitting, therefore the dimers were placed manually and interconnected as proposed in 26. d) Cartoon depicting ClpXP1/2 dimerization via the ZBD domains of two opposing ClpX hexamers. Arrows indicate the flexibility of the complex.