Figure 2. Cryo-EM structure of the ClpXP1/2 protein degradation machinery.
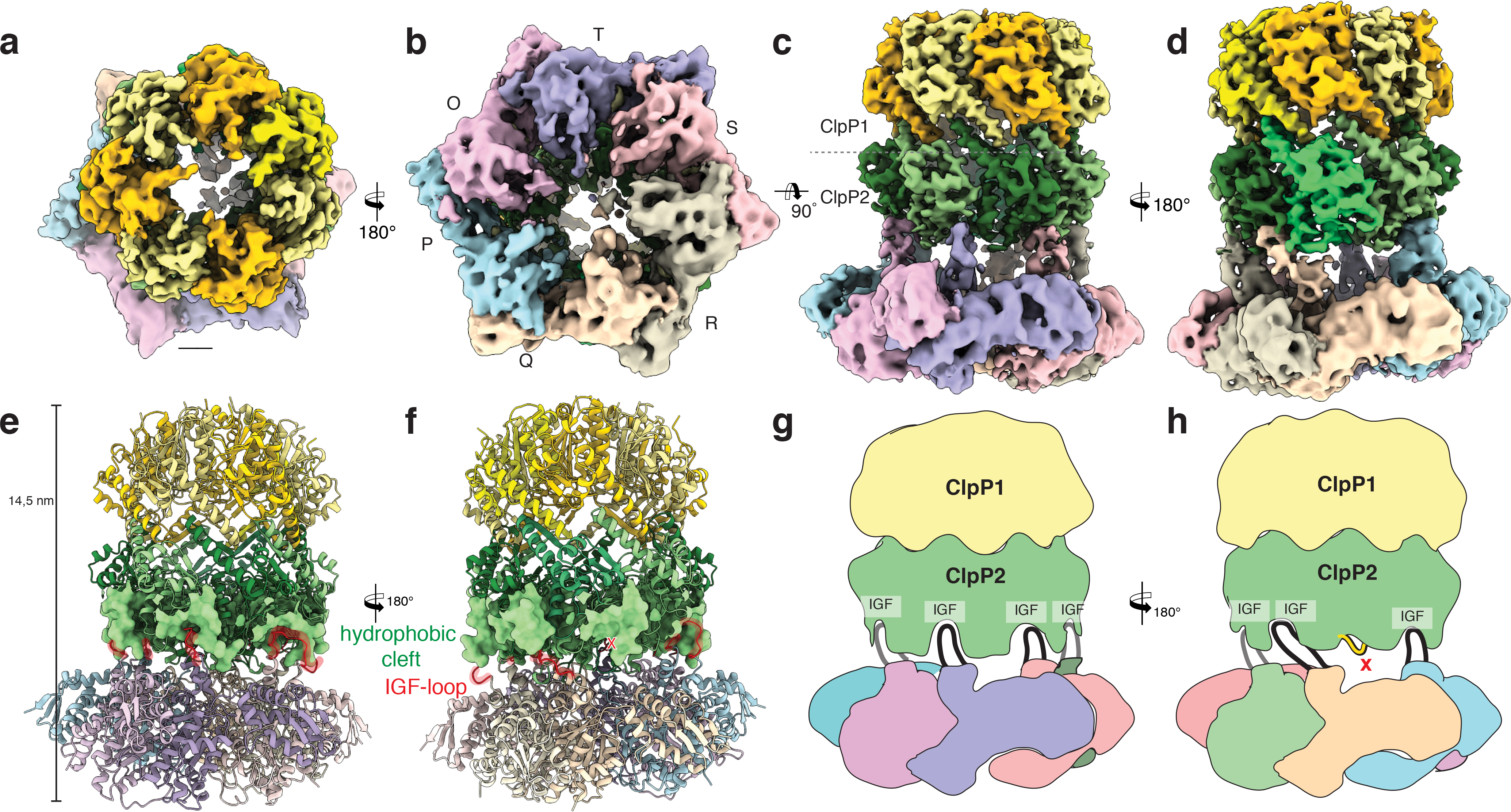
a-d) Cryo-EM density of ClpXP1/2 shown from the top (a), bottom (b) and side (c,d). ClpP1 and ClpP2 subunits are colored in khaki, orange and dark, light green, respectively. ClpP2 subunit J is highlighted in mint green. Note that this is the only ClpP2 subunit not interacting with ClpX via an IGF-loop. Each subunit of ClpX is assigned a different color. This color code is maintained throughout the manuscript. e-f) Molecular model of ClpXP. The hydrophobic pockets of ClpP2, each spanning two ClpP2 subunits, are shown as surface. The IGF interaction loops are highlighted in red. g-h) Cartoon depicting how the ClpX hexamer interacts with the ClpP2 heptamer via the six IGF-loops. Note the extended conformation of IGF-loop of ClpX subunit Q.
