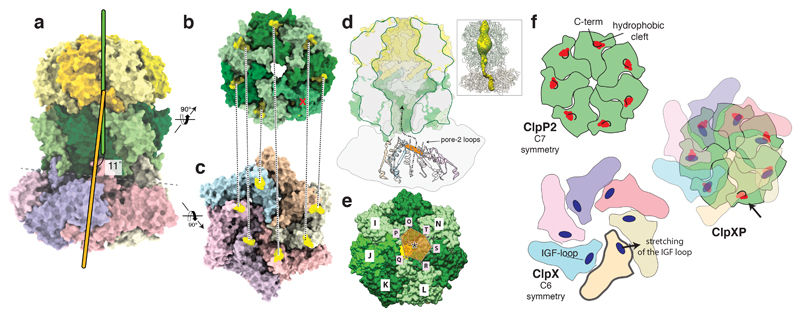
Figure 3. Symmetry mismatch between ClpP1/2 and ClpX.
a) Molecular model of ClpXP1/2. The symmetry axes of the ClpP1/2 and ClpX are shown in green and orange, respectively. b-c) The ClpP2 heptamer (b) and the ClpX hexamer (c) are shown from the bottom and the top, respectively, perpendicular to the plane of the ClpP2-ClpX interface. The positions of the IGF-loops and the hydrophobic grooves are highlighted in yellow and connected by dashed lines. d) Cut-away view of the ClpP density. Secondary structure elements directly prior (residues 170-189) and after the pore-2-loops (residues 202-220) of ClpX are shown in ribbon representation. The pore-2-loops are not resolved in the cryo-EM density and not shown here. In order to indicate the arrangement and positioning of the pore-2-loops, as well as the position of the upper opening of the ClpX channel relative to the ClpP2 pore, a plane was calculated using the Cα atoms of Gly202 as anchor points and depicted here in orange. The plane is tilted and shifted relative to the ClpP channel axis, suggesting a spiral staircase-like arrangement of the pore-2-loops. The dashed line with the arrowhead indicates the pathway of substrate translocation from ClpX towards the ClpP proteolytic chamber. The inset shows the skin surface of the ClpXP pore. e) Molecular surface of ClpP2 shown from the bottom. Rosetta models of the pore-2-loops of ClpX are shown as ribbons. The black star indicates the positioning of the ClpX channel opening relative to the ClpP channel opening (yellow star). f) Schematic model of the ClpX-ClpP2-binding mechanism. Left images depict axial views of the ClpP2 heptamer (green) and the ClpX hexamer prior assembly of the ClpXP protease. The main interaction elements, the ClpX IGF-loops and ClpP2 hydrophobic grooves are highlighted. The remaining “free” ClpP2 hydrophobic groove stays shielded by the respective C-terminus (arrow).