Figure 7. ClpX binds to ClpP in a similar manner like ADEP, but does not induce ClpP pore widening.
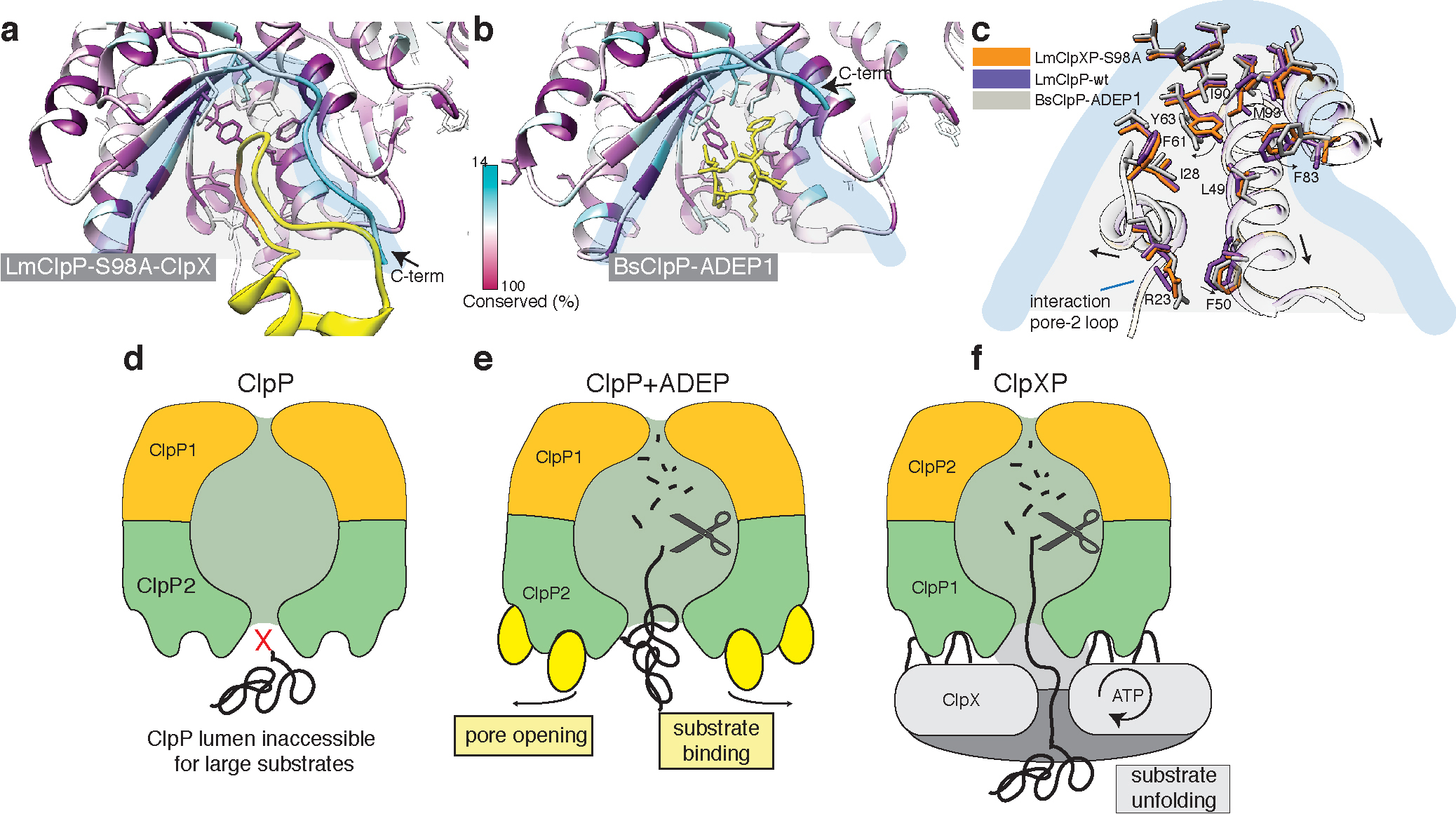
a) Local molecular interactions at one of the seven binding pockets between ClpP and the IGF-loop of ClpX. Residues of ClpP are colored by sequence conservation. The IGF-loop is shown in yellow with the IGF residues highlighted in orange. b) Interface of ADEP2 (yellow) with BsClpP (PDB 3KTI) (colored by conservation). c) Structural superposition of the binding pockets of ClpX-bound LmClpP2-S98A, ADEP1-bound BsClpP (PDB 3KTI) and “free” LmClpP2 (PDB 4RYF). Arrows indicate changes upon ADEP binding. d-f) Regulation of ClpP by ClpX and ADEP. The central pore of the ClpP protease is closed and entry of folded proteins into the proteolytic chamber is not allowed (d). ADEP binding to the binding pockets of ClpP induces pore opening. The proteolytic chamber is now accessible for unfolded proteins, leading to unregulated protein degradation and cell death. (e) ClpX binds in the same hydrophobic pockets on ClpP but does not induce pore opening. ClpP and ClpX form a continuous pore instead, with ClpX unfolding target proteins and forwarding them to the proteolytic chamber of ClpP for degradation in a regulated manner (f).
