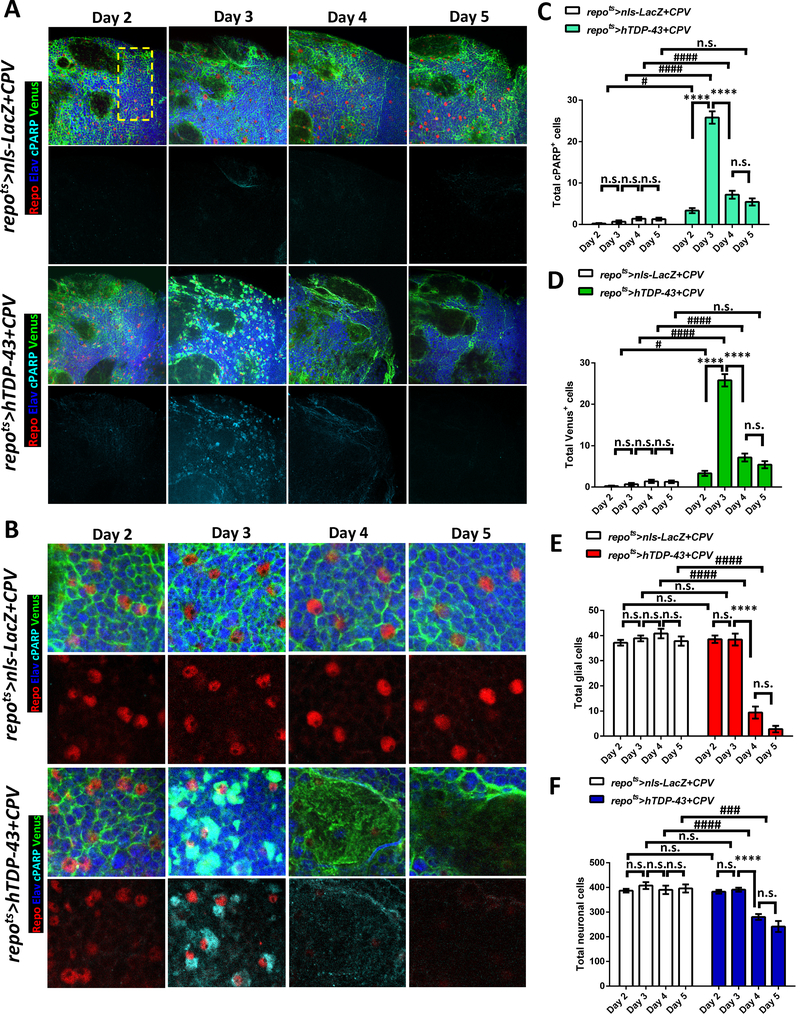
Figure 2. Induction of hTDP-43 in Glia Activates Caspase-3 and Causes Glial and Neuronal Cell Loss.
(A) CPV reporter was used to trace Caspase-3 activity after induction of nls-LacZ (repots>nls-LacZ+CPV) vs hTDP-43 (repots>hTDP-43+CPV) in glia over a time-course post induction (Day 2 – Day 5). The glia and neurons were independently labeled with cell type specific markers, Repo (Red) and Elav (Blue), and the Venus and cPARP reporters from CPV are shown with Green and Cyan. Cells located within the region highlighted with a dashed yellow rectangle were quantified. The total numbers of each marker are shown in (C-F). Scale bar=20 μm. See also Figure S3.
(B) Higher magnification from (A). Scale bar=10 μm.
(C) Total numbers of cPARP positive cells from repots>nls-LacZ+CPV and repots>hTDP-43+CPV were quantified. cPARP positive numbers from each time-point are shown (mean ± SEM). Average cPARP numbers in repots>nls-LacZ+CPV were 0.1±0.1 (Day 2, n=10), 0.9±0.4 (Day 3, n=12), 1.3±0.3 (Day 4, n=13) and 1.3±0.4 (Day 5, n=7). cPARP numbers in repots>hTDP-43+CPV were 3.8±0.7 (Day 2, n=12), 24.6±1.3 (Day 3, n=11), 7.6±1.1 (Day 4, n=12) and 5.4±0.9 (Day 5, n=7). ****p<0.0001, n.s., no significant difference (One-way ANOVA). #p<0.05, ####p<0.0001, n.s., no significant difference (Two-way ANOVA).
(D) Total numbers of Venus positive cells from repots>nls-LacZ+CPV and repots>hTDP-43+CPV. Venus were quantified (mean ± SEM) for each time-point. Average numbers of Venus positive cells in repots>nls-LacZ+CPV were 0.1±0.1 (Day 2, n=10), 0.9±0.4 (Day 3, n=12), 1.3±0.3 (Day 4, n=13) and 1.3±0.4 (Day 5, n=7). Venus numbers in repots>hTDP-43+CPV were 3.8±0.7 (Day 2, n=12), 24.6±1.3 (Day 3, n=11), 7.6±1.1 (Day 4, n=12) and 5.4±0.9 (Day 5, n=7). ****p<0.0001, n.s., no significant difference (One-way ANOVA). #p<0.05, ####p<0.0001, n.s., no significant difference (Two-way ANOVA).
(E) Total numbers of glia labeled with the pan-glial marker, Repo are shown from repots>nls-LacZ+CPV and repots>hTDP-43+CPV at different time points. Average numbers of Repo-labeled glia numbers are shown (mean ± SEM). Glial numbers in repots>nls-LacZ+CPV were 37.0±1.2 (Day 2, n=10), 39.5±1.3 (Day 3, n=12), 43.3±2.1 (Day 4, n=13) and 37.9±1.8 (Day 5, n=7). Glial numbers in repots>hTDP-43+CPV were 40.0±1.5 (Day 2, n=12), 38.8±2.7 (Day 3, n=11), 9.4±2.8 (Day 4, n=12) and 2.9±1.3 (Day 5, n=7). ****p<0.0001, n.s., no significant difference (One-way ANOVA). ####p<0.0001, n.s., no significant difference (Two-way ANOVA).
(F) Total numbers of neurons (mean ± SEM) labeled with the pan-neuronal marker, Elav, are shown from repots>nls-LacZ+CPV and repots>hTDP-43+CPV during over a time-course. Total neuronal numbers in repots>nls-LacZ+CPV were 384.8±8.1 (Day 2, n=10), 413.9±16.5 (Day 3, n=12), 396.1±21.4 (Day 4, n=13) and 359.9±16.6 (Day 5, n=7). Neuronal numbers in repots>hTDP-43+CPV were 379.4±8.0 (Day 2, n=12), 389.4±5.7 (Day 3, n=11), 274.0±12.9 (Day 4, n=12) and 241.6±22.2 (Day 5, n=7). ****p<0.0001, n.s., no significant difference (One-way ANOVA). ###p<0.001, ####p<0.0001, n.s., no significant difference (Two-way ANOVA).