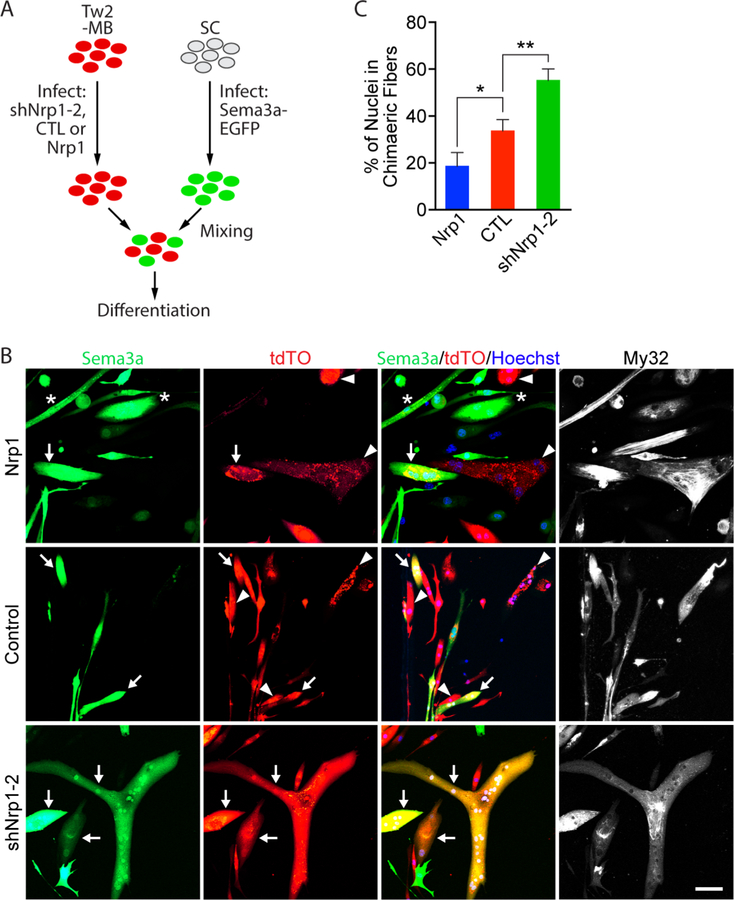
Figure 6. The Nrp1-Sema3a signaling axis represses chimeric fusion between Tw2-MB and Sema3a over-expressing primary myoblasts.
(A) Experimental scheme for chimeric fusion assay. Tw2-MB were infected with retroviruses expressing shNrp1–2, control empty vector, or Nrp1, and mixed with primary myoblasts (SCs) infected with retroviruses expressing Sema3a-EGFP in equal numbers. Cells were then differentiated for 7 days. (B) SCs over-expressing Sema3a (green) were mixed with Tw2-MB (tdTO+) infected with Nrp1 (top), control empty vector (middle), or shNrp1–2 (bottom) retroviruses and differentiated for 7 days. Cells were fixed and stained with Hoechst (blue) and an antibody against fast myosin (MY32; white). Arrow indicates chimeric myotubes that are both Sema3a+ and tdTO+; * represents myotubes that Sema3a+ only; and arrowhead represents myotubes that are tdTO+ only. Scale bar: 50 µm. (C) Quantification of percent of nuclei in chimeric fibers for Figure 6B. Percent of nuclei in chimeric fibers was calculated as the percent of the number of nuclei in chimeric fibers over the total number of nuclei (red-only, green-only, and chimeric myofibers). Three fields per sample per experiment were quantified. Three separate experiments were performed. *: p < 0.05, **: p < 0.005. See also Figure S6. Source data for 6C is provided in Supplementary Table 1.