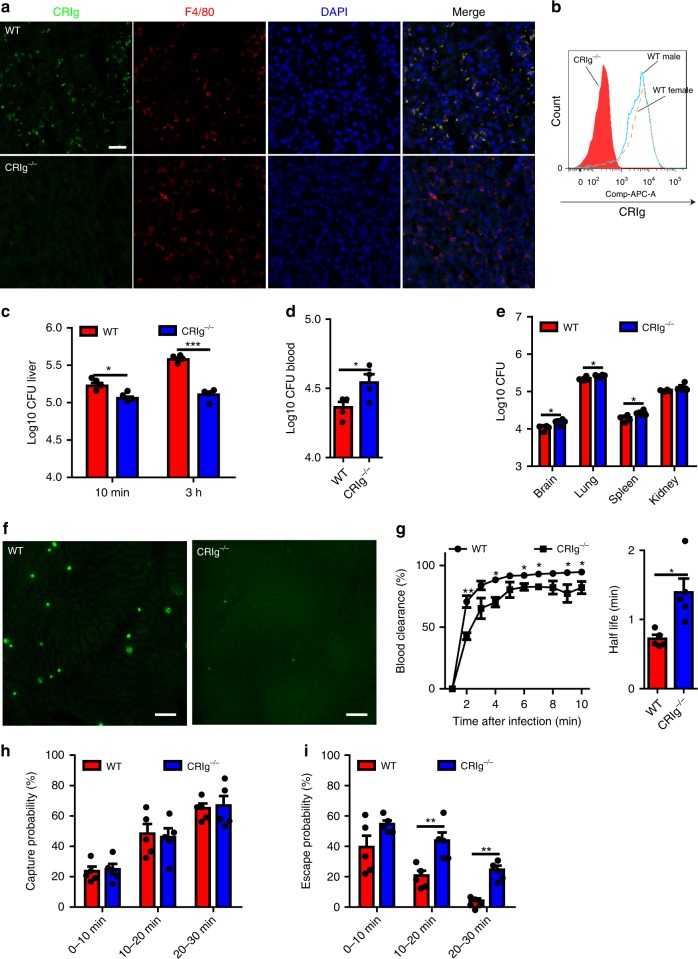
Fig. 4.
CRIg is the receptor on KCs mediating the capture of C. neoformans. a Immunofluorescence staining showing the expression of CRIg (green) on KCs (red) of WT mice (upper panel) but not CRIg−/− mice (lower panel). The nuclei were stained with DAPI (blue). b The expression of CRIg on KCs of WT (male and female) but not CRIg−/− mice (gated on CD45+F4/80+CD11bint population). c Liver CFU was enumerated in WT and CRIg−/− mice (n = 4 mice/group) 10 min and 3 h after i.v. infection with 5 × 106 C. neoformans. d Blood CFU was enumerated in WT and CRIg−/− mice (n = 4 mice/group) 10 min post i.v. infection of 5 × 106 C. neoformans. e CFU of different organs was enumerated in WT and CRIg−/− mice (n = 4 mice/group) 3 h after i.v. infection of 5 × 106 C. neoformans. f Representative IVM images showing yeast cells (green) captured in the liver of WT and CRIg−/− mice (n = 5 mice/group) 30 min after i.v. infection with 100 × 106 GFP-labeled C. neoformans. g Vascular clearance and half-life of circulating C. neoformans at various time points after injection of 100 × 106 yeast cells into tail vein of WT and CRIg−/− mice (n = 5 mice/group) by analysis of IVM videos. h The possibility of circulating C. neoformans being captured was calculated by analysis of IVM videos from infected WT and CRIg−/− mice (n = 5 mice/group). i The possibility of trapped C. neoformans being released back to circulation was calculated by analysis of IVM videos from infected WT and CRIg−/− mice (n = 5 mice/group). Scale bars: 25 µm. Data are expressed as mean ± SEM of two independent experiments. All data are from biologically distinct samples. *p < 0.05, **p < 0.01, ***p < 0.001. p values were calculated via two-way ANOVA (c, h, i) or Student’s t test (d, e, g). Source data are provided as a Source Data file