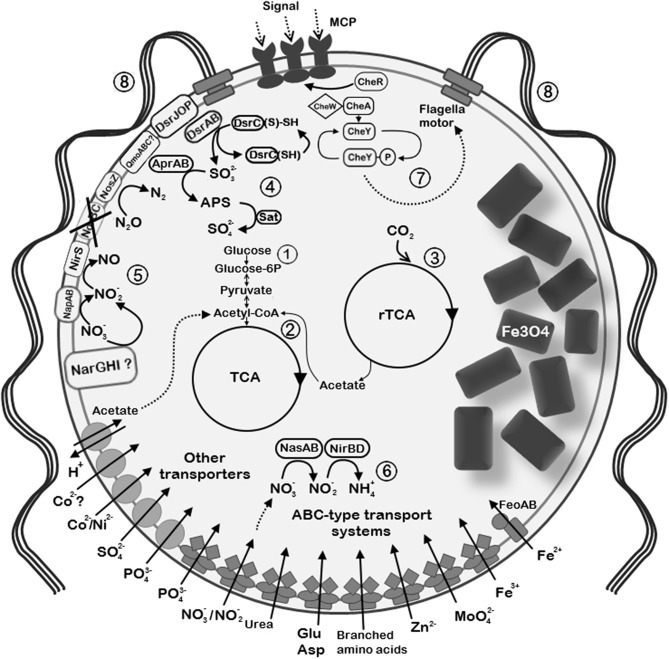
Figure 7.
Reconstruction of the selected metabolic and structural features of “Ca. Magnetaquicoccus inordinatus” based on genome analysis and TEM micrographs. The following metabolic pathways are emphasized: (1) glycolysis and glyconeogenesis; (2) tricarboxylic acid cycle (TCA); (3) autotrophic CO2 fixation through reverse TCA (rTCA); (4) sulfide oxidation trough reverse Dsr (dissimilatory sulfite reductase) pathway. Sulfite which is formed via Dsr operation is further oxidized to sulfate through the intermediate 5'-adenylyl sulfate (APS) by APS reductase (AprAB) and sulfate adenylyltransferase (Sat). (5) Dissimilatory nitrate reduction is possible through the periplasmic nitrate reductase (NapAB) and putative membrane-bound nitrate reductase complex (NarGHI). Nitrite reduction is further possible by nitrite reductase NirS. Since norBC genes were not found (crossed out), might be not capable of nitric oxide reduction, whereas nitrous oxide can be reduced to nitrogen by nitrous oxide reductase (NosZ). (6) Assimilatory nitrate and nitrite reduction in the cytoplasm through nitrate reductase (NasA) and nitrite reductase NirBD, respectively. (7) Chemotaxis complex is represented by methyl-accepting chemotaxis proteins (MCP) and Che proteins. (8) The species presumably synthesizes multiple flagella due to the presence of high number of flagellin genes. Since two bunches of flagella are common among magnetotactic cocci, similar organization is assumed in “Ca. Magnetaquicoccus inordinatus”.