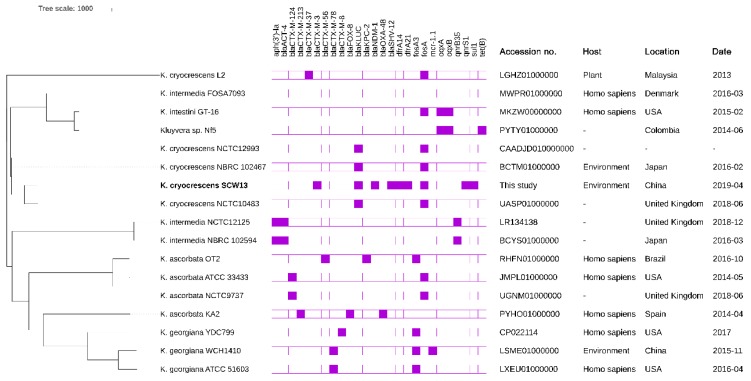
Figure 3.
A phylogenetic analysis of the core genomes of K. cryocrescens strain SCW13 identified in this study (marked in bold) and 15 Kluyvera genomes deposited in the GenBank database (last accessed July 12, 2019). From left to right: (1) A maximum likelihood tree of Kluyvera spp strains. The phylogeny was inferred from the recombination-filtered single nucleotide polymorphism (SNP) alignment obtained by aligning a genome of Kluyvera isolate against the complete genome of K. cryocrescens NBRC 102467. (2) A heatmap of the antimicrobial resistance genes as determined by ABRicate. The presence or absence of antibiotic resistance genes is indicated by filled or empty squares, respectively. (3) The annotation of each Kluyvera isolate, including GenBank accession no., hosts of isolates, locations, and collection dates. -, not available.