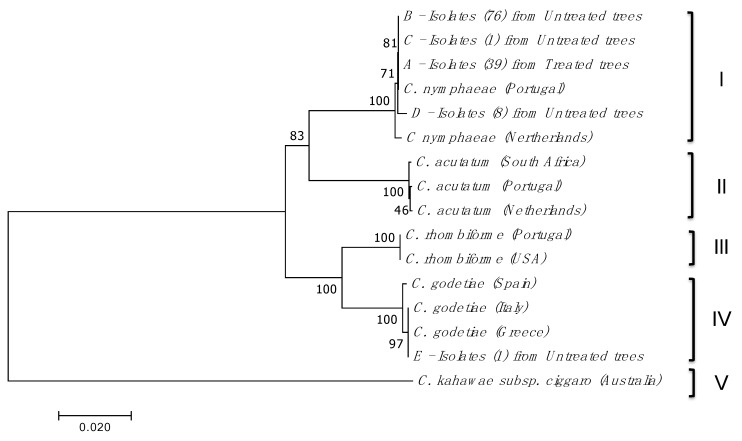
Figure 2.
The NJ analysis was constructed from the sequence alignment of tub2, GAPDH, ACT, CHS-1 and HIS-3 (except for C. gloeosporioides complex) partial genes from isolates from C. acutatum complex (C. nymphaeae, C. godetiae, C. acutatum and C. rhombiforme) and C. gloeosporioides complex (C. kahawae subsp. ciggaro), using 125 isolates obtained in this study plus 11 sequences retrieved from the GenBank database, totalling 133 sequences. Repeated sequences within each group of isolates were omitted. Each sequence variant was named with a letter (A to E; except for A and B which are identical but differ in the management regime), following the number of isolates within each group. NJ analysis included 16 sequences. Multiple sequence alignments were generated using MEGA 7 and the neighbour joining (BioNJ algorithms), based on calculations from pairwise nucleotide (nt) sequence distances for gene nt analysis. Numbers above the lines indicate bootstrap scores out of 1000 replicates.