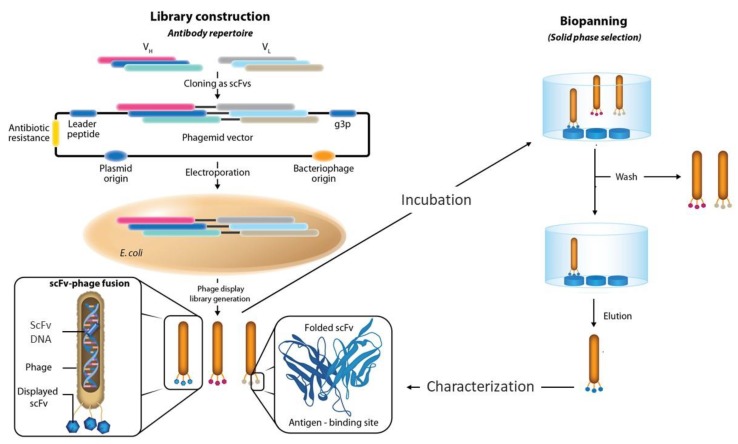
Figure 1.
Phage display methodology. The method consists of building a library (left panel) of peptide or protein variants—or in the case of antibodies, an antibody gene repertoire—and affinity selecting (right panel) specific antibody-phage fusions via affinity with the target of interest. An antibody library is commonly built in a phagemid vector as fusions to one of the phage coat proteins. The repertoire of antibody variants, for instance cloned in the scFv format, is used to transform E. coli via electroporation and is expressed together with the other viral components. The population of specific scFvs-phage particles is enriched via binding to the target of interest. Non-specific or not-well-folded scFvs are removed via washing steps. The specific scFv-phage fusions are then eluted using diverse elution methods. The eluted phage bearing specific scFv genes can be used to infect E. coli and thus amplify the population of specific antibodies via additional rounds of selections. Alternatively, the outcome of the selection is characterized in the screening phase.