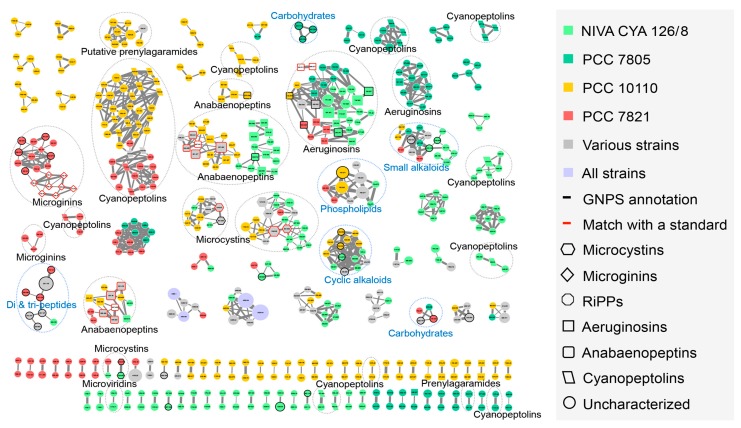
Figure 1.
Molecular network generated from MS/MS spectra of the four Planktothrix strains using Global Natural Products Social Molecular Networking (GNPS) tool (all data and results are freely available on the GNPS server at the address http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=98e54f0fa2a84efeb82efa0d24e4d974). The GNPS algorithm compares all MS/MS spectra by aligning them one by one, grouping identical molecules (presenting identical mass and fragmentation pattern) and assigning a cosine score ranking from 0 to 1 to each alignment, allowing network reconstruction of the link between each molecule according to the cosine score calculated between all molecules (cosine score significance threshold set to 0.6). Analytes whose individual masses match with known secondary metabolites from cyanobacteria are indicated with specific shapes. Correspondences with standard molecules from cyanobacteria similarly analyzed or with components from the fragmentation pattern library available from the GNPS server are indicated by heavy red and black lines, respectively. Only clusters of at least two nodes are represented.