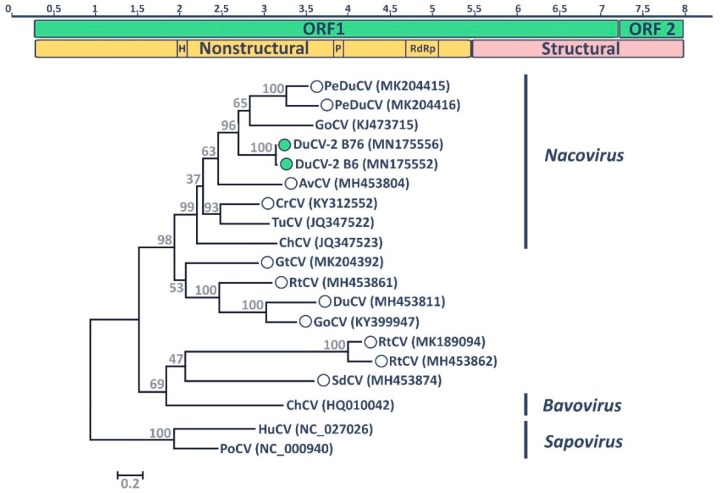
Figure 3.
Molecular characteristics of DuCV-2 B6. The genome organization of the novel virus is depicted at the top. The two identified ORFs are illustrated by green rectangles, while nonstructural and structural protein regions are indicated in yellow and pink, respectively. Enzymatic domains typical of caliciviruses are indicated (H: helicase/NTPase; P: 3C-like cysteine protease; RdRp: RNA-dependent RNA polymerase) and the scale bar above indicates genomic position in Kb. The phylogenetic placement of DuCVs-2 B6 and B76 within the genus Nacovirus based on the predicted amino acid sequence of the major capsid protein (VP1) is shown at the bottom. Hosts of viruses used for phylogenetic reconstruction are indicated within the strain designation (PeDu: Pink-eared duck; Du: Duck; Go: Goose; Av: Avocet; Cr: Crane; Tu: Turkey; Ch: Chicken; Gt: Grey teal; Rt: Ruddy turnstone; Sd: Shelduck; Hu: Human; Po: Porcine), while accession numbers of sequences are indicated in parentheses. Viral genera are depicted on the right and the two mammalian caliciviruses of the genus Sapovirus were used as an outgroup. Recently identified viruses that lack official taxonomic designation are indicated by circles and the viruses identified in this study are indicated by filled green circles. The tree was built with the maximum likelihood method [47] using MEGA 7 [43] based on the Le Gascuel model [50], identified as the best-fitting model by the model test in MEGA. A discrete Gamma distribution was used to model evolutionary rate differences among sites, branch lengths are proportional to genetic distances as indicated by the scale bar, and the outcome of the bootstrap analysis [48] is shown next to the nodes.