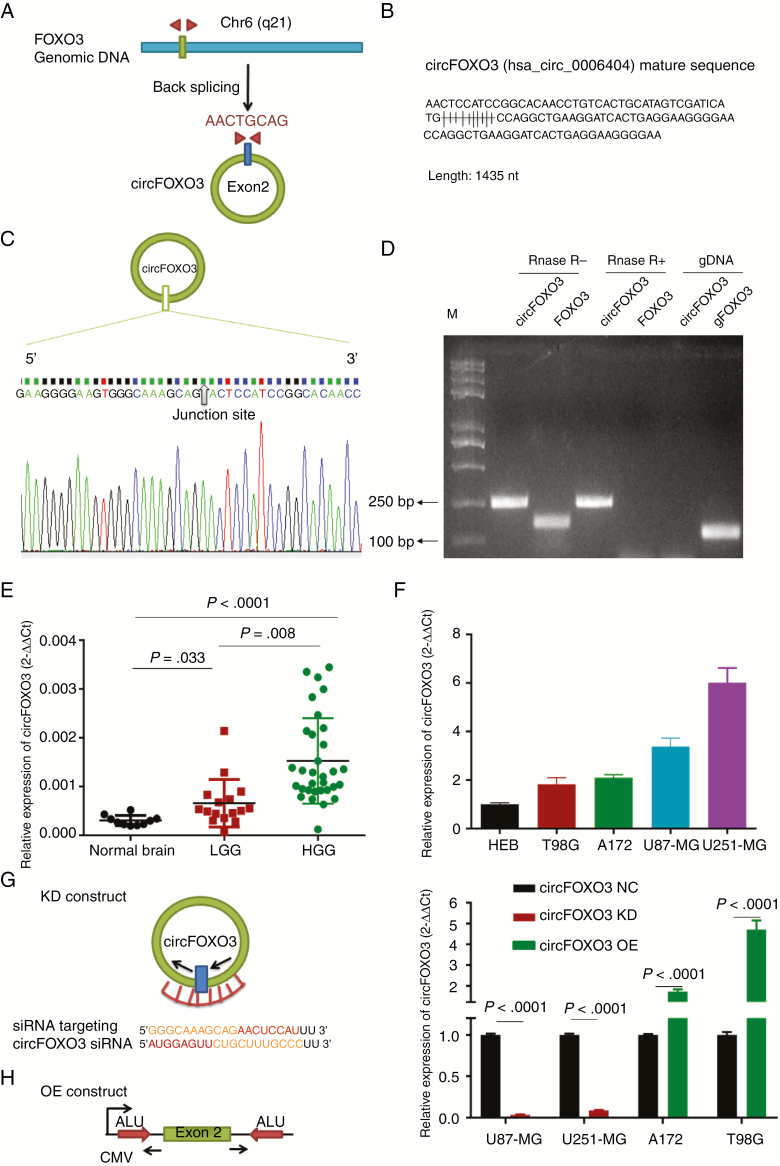
Fig. 1.
Identification and expression of circFOXO3 in GBM tissues and cells. (A) The structure and part of the sequence of the junction of circFOXO3 are provided, and divergent (red) primers were designed to amplify the back-splicing products. (B) Part of the mature circFOXO3 sequence. (C) Sanger sequencing after PCR using the indicated divergent flanking primers confirmed the “head-to-tail” splicing of circFOXO3. (D) Total RNA from cells with or without RNase R treatment was subjected to PCR. (E) CircFOXO3 transcript levels were significantly higher in HGG than in LGG (LGG vs normal controls: P = 0.033, HGG vs LGG, P = 0.008; one-way, P < 0.0001). (F) Quantitative RT-PCR analysis of circFOXO3 in GBM cells and HEBs. (G) A sketch of the short hairpin circFOXO3 vector structure. (H) A sketch of the circFOXO3 overexpression vector structure. ALU: Complementary ALU pairs were identified as at least one plus strand and one minus strand ALU element on opposite sides of the back-splice, which was required for the formation of circRNA; CMV: a common promoter in vectors. (I) Quantitative RT-PCR analysis of circFOXO3 expression in GBM cells. A multiple comparisons test adjusted P-value of <0.05 was considered statistically significant. Error bars represent the mean ± SD.