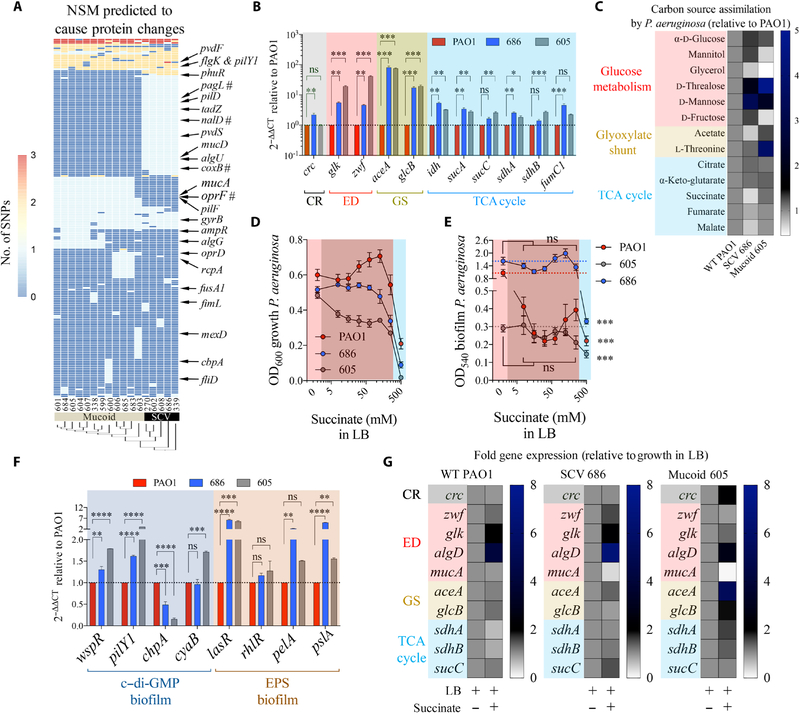
Fig. 4. Metabolism of P. aeruginosa isolates is consistent with adaptation to succinate.
(A) Nonsynonymous mutations (NSM) in 17 P. aeruginosa isolates (SCV and mucoid variants) compared with PAO1 control. #: stop codon mutations. (B) Fold mRNA expression relative to PAO1 for selected metabolic genes in SCV 686 and mucoid 605 (n = 3). (C) Relative single-carbon source assimilation for PAO1, SCV 686, and mucoid 605 strains (n = 6). (D) Bacterial final point growth in increasing concentrations of succinate in LB (n = 3). (E) Bacterial biofilm in increasing concentrations of succinate in LB (n = 3). (F) Relative mRNA expression against control PAO1 by quantitative reverse transcription polymerase chain reaction (qRT-PCR) of genes that regulate c-di-GMP and EPS required to produce biofilm (n = 3). (G) Regulation of metabolic genes by succinate in P. aeruginosa SCV 686 and mucoid 605 clinical isolates (n = 3). Data are shown as means ± SEM. (B and F) One-way ANOVA; (D and E) two-way ANOVA. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.