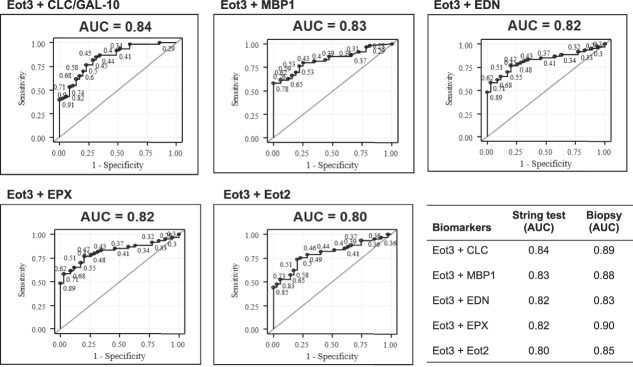
Figure 5.
ROC curves distinguish subjects with active EoE (≥15 Eos/hpf) from inactive EoE (<15 Eos/hpf). ROC analyses used the level of Eot3 plus one additional biomarker captured by the 1-hour EST or measured in mucosal biopsy extracts (lower right panel; see Figure S3, Supplementary Digital Content 1, http://links.lww.com/AJG/B281). Results are shown for Eot3, Eot3 plus CLC (Gal-10), Charcot-Leyden crystal protein/galectin-10; MBP-1, major basic protein-1; EPX, eosinophil peroxidase; EDN, eosinophil-derived neurotoxin; and Eot2, eotaxin 2. Points on the ROC curves are labeled by their predictive probabilities. The AUC is indicated above each panel; AUC values >0.80 are considered highly predictive. The table (lower right panel) shows the comparative AUC values for the 1-hour EST vs mucosal biopsy extracts for the indicated biomarker combinations. AUC, area under the curve; CLC/GAL-10, Charcot-Leyden crystal protein/Galectin-10; EoE, eosinophilic esophagitis; EDN, eosinophil-derived neurotoxin; Eot2, eotaxin 2; Eot3, eotaxin 3; EPX, eosinophil peroxidase; MBP-1, eosinophil granule major basic protein 1; ROC, receiver operating characteristic.