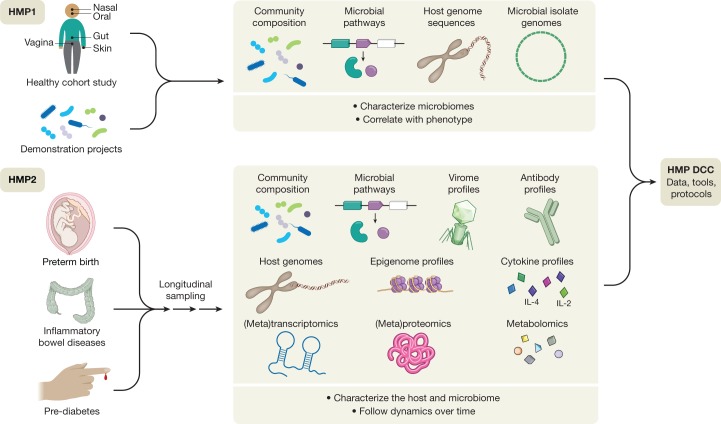
Fig. 1. The first and second phases of the NIH Human Microbiome Project.
The ten-year NIH Human Microbiome Project (HMP) program, organized into two phases (HMP1 and HMP2), developed reference sequences, multi-omic data sets, computational and statistical tools, and analytical and clinical protocols as resources for the broader research community. The HMP1 focused on the characterization of microbial communities from numerous body sites (oral, nasal, vaginal, gut, and skin) in a baseline study of healthy adult subjects, and included a set of demonstration projects that focused on specific diseases or disorders. The HMP2 expanded the repertoire of biological properties analysed for both host and microbiome in three longitudinal cohort studies of representative microbiome-associated conditions: pregnancy and preterm birth (vaginal microbiomes of pregnant women), inflammatory bowel diseases (gut microbiome) and prediabetes (gut and nasal microbiomes). These studies followed the dynamics of these conditions through multi-omic analyses of multiple measurement types over time, including changes in microbial community composition, viromics, metabolomic profiles, gene expression and protein profiles from both host and microbiome, and host-specific properties such as genetic, epigenomic, antibody, and cytokine profiles, along with other study-specific features. All sequences and multi-omic data, clinical information, and tools from both HMP1 and HMP2 are housed in the HMP Data Coordination Center (DCC) or referenced public or controlled-access repositories to serve as a central resource for the research community.