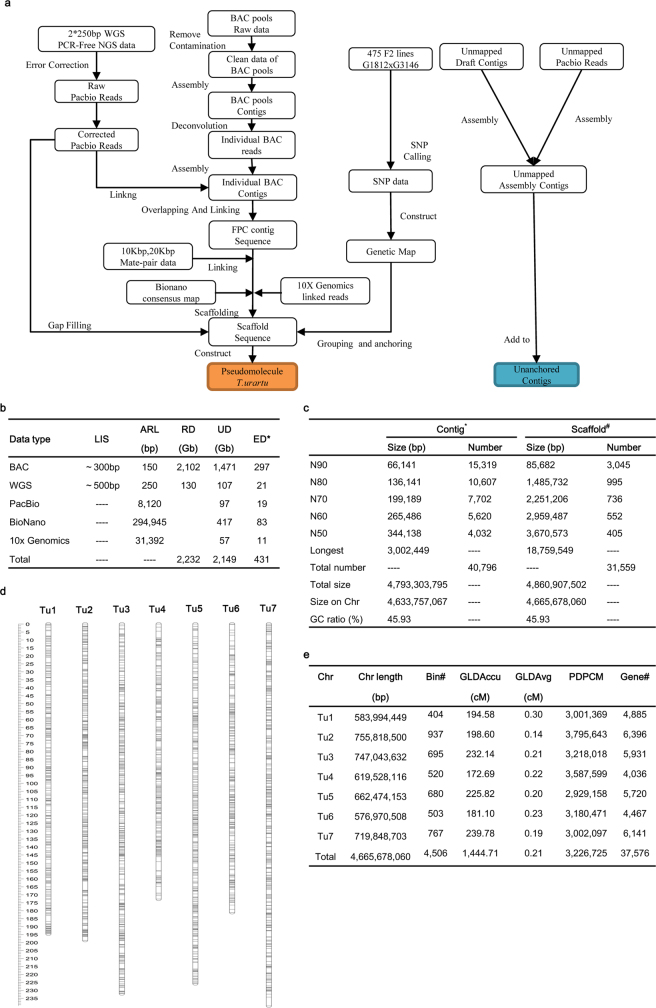
Extended Data Fig. 1. T. urartu genome assembly.
a, Schematic workflow for genome sequencing, assembly and chromosomal assignment with a high-density SNP map. b, Statistics of sequencing data. *Calculated from the estimated genome size of 4.94 Gb. LIS, library insert size; ARL, average read length; RD, raw data; UD, usable data; ED, effective depth. c, Summary of the Tu genome assembly. *Contig: contiguous sequence without Ns assembled with Illumina reads, corrected by PacBio reads. #Scaffold: Sequence with Ns, in which two or more contigs were connected by mate-pair reads, BioNano genome maps and 10× Genomics linked reads. d, High-resolution genetic map of T. urartu using SNP markers. The SNP markers were identified from an F2 population (475 individuals) derived from a cross between accessions G1812 and G3146 of Tu. e, Summary of physical length and genetic map of seven pseudomolecules. Chr, chromosome; GLDAccu, genetic linkage distance accumulation; GLDAvg, genetic linkage distance on average; PDPCM, physical distance per centiMorgen (cM).