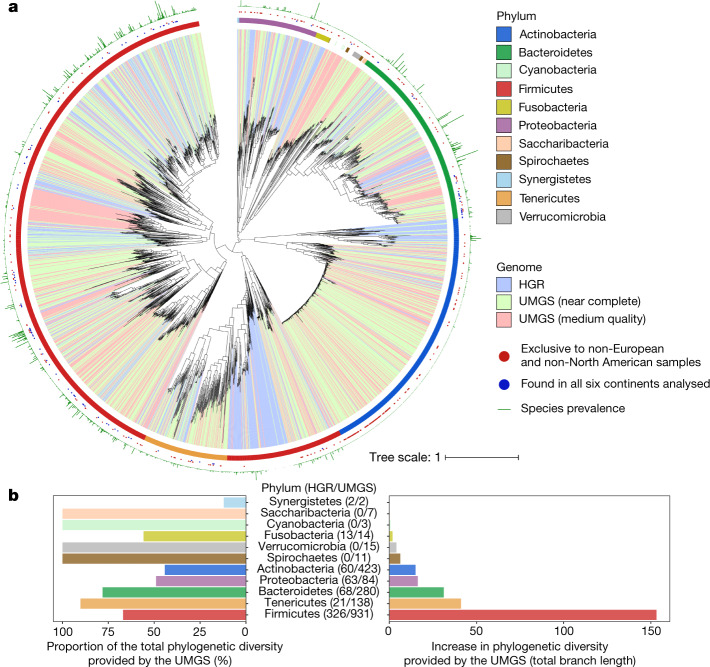
Fig. 3. Phylogeny of reference and uncultured human gut bacterial genomes.
a, Maximum-likelihood phylogenetic tree comprising the 553 genomes belonging to the HGR, and 1,952 to UMGS. Clades are labelled according to genome type (HGR, near-complete or medium-quality UMGS) and the corresponding phylum is depicted in the first outer layer. Blue and red dots in the second layer denote genomes that were found in at least one sample from all six continents analysed (Africa, Asia, Europe, North America, South America and Oceania), or exclusively detected in non-European, non-North American samples, respectively. Green bars in the outermost layer represent the prevalence of the genome among the 13,133 metagenomic datasets. b, Level of increase in phylogenetic diversity provided by the UMGS, relative to the complete diversity per phylum (left) and represented as absolute total branch lengths (right). The number of HGR and UMGS genomes assigned to each phylum is depicted in brackets (HGR/UMGS).