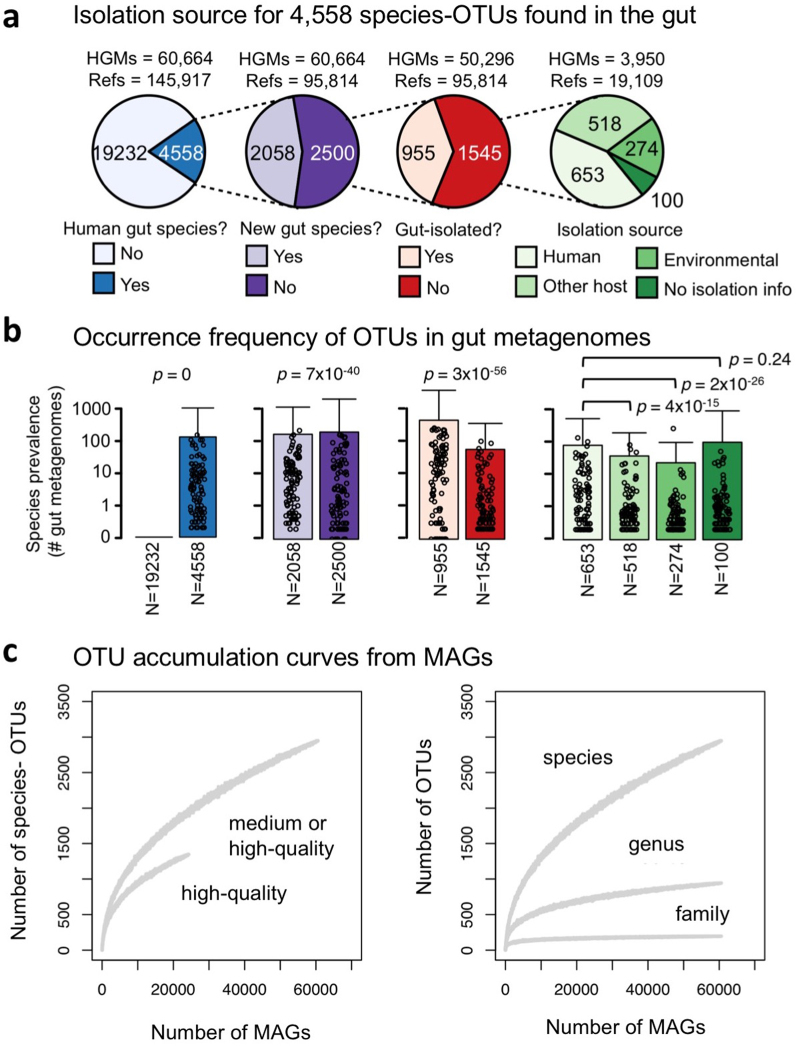
Extended Data Fig. 6. Annotation and accumulation of human gut OTUs.
a, Of the 23,790 species-level OTUs identified from MAGs and reference genomes, 4,558 were classified as being from the human gut on the basis of (1) having a MAG from the HGM dataset, (2) being detected in a human gut metagenome via read-mapping with IGGsearch or (3) containing a reference genome with metadata that indicate isolation from a human stool sample. Of the 4,558 gut OTUs, 2,058 are represented exclusively by MAGs from the current study and are therefore newly identified. Of the remaining 2,500 represented by reference genomes, only 955 contained a gut-isolated reference genome. The remaining 1,545 OTUs either lack isolation metadata or contain metadata that indicate other isolation sources, including human, non-human and environmental. For example, several gut species from non-host-associated environments were isolated from human food products, including milk, cheese, meat and fermented foods. b, The occurrence frequency of all 4,558 gut OTUs was estimated across 3,810 human stool metagenomes using IGGsearch. For bar plots, the centre bar indicates the mean, the error bar indicates the standard deviation and 100 random data points are overlaid. P values are from two-sided Wilcoxon rank-sum tests. c, Accumulation curves that indicate that the discovery of genus- and family-level OTUs from MAGs has saturated, but that the discovery of species-level OTUs has not. To make the plots, MAGs were randomly sampled without replacement, and the number of unique OTUs was counted for each sample.