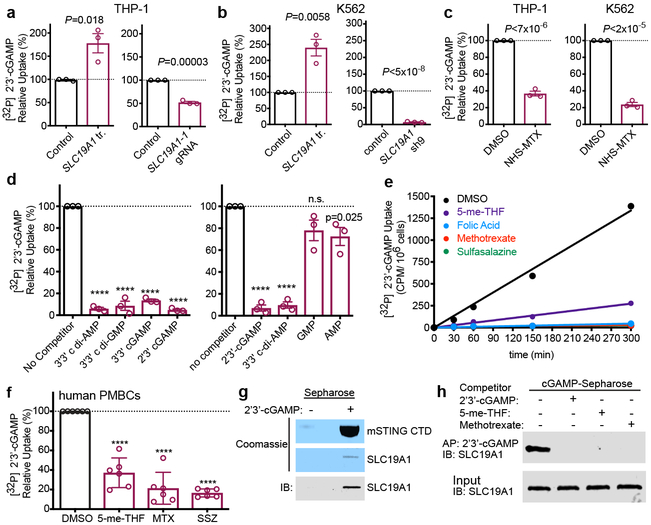
Figure 4.
SLC19A1 transports CDNs. a, Normalized [32P] 2’3’-cGAMP uptake after one hour by THP-1 monocytes transduced with empty vector (control) or SLC19A1 expression vector (left panel), or transduced with a non-targeting control CRISPRi gRNA or SLC19A1 CRISPRi gRNA (right panel). b, Normalized [32P] 2’3’-cGAMP uptake after one hour by K562 cells transduced with empty vector (control) or SLC19A1 expression vector (left panel), or transduced with a non-targeting control shRNA (control) or SLC19A1 shRNA (right panel). c, Normalized [32P] 2’3’-cGAMP uptake after one hour in DMSO or NHS-methotrexate (MTX) (5 μM) treated THP-1 (left panel) and K562 (right panel) cells. d, Normalized [32P] 2’3’-cGAMP uptake after one hour by THP-1 monocytes in the presence and absence of 100 μM competing, unlabeled cyclic dinucleotides (left panel) or 200 μM competing, unlabeled nucleotides (right panel). e, Time course of [32P] 2’3’-cGAMP uptake in THP-1 monocytes in the presence and absence of 500 μM competing, unlabeled (anti-) folates and sulfasalazine. f, Normalized [32P] 2’3’-cGAMP uptake after three hours in human PBMCs from six healthy donors in the presence and absence of 500 μM competing, unlabeled (anti-) folates and sulfasalazine. g, Coomassie staining and western blot analysis of pulldowns by 2’3’-cGAMP (+) or control (−) Sepharose of mSTING-CTD or hSLC19A1. h, Western blot analysis of hSLC19A1 affinity purification (AP) with 2’3’-cGAMP Sepharose in the absence (−) or presence (+) of free, unbound 2’3’-cGAMP, 5-me-THF, or methotrexate (250 μM). In panels a-d, data are means ± SEM of n=3 biological replicates. In panel e, data are means ± SD of n=3 technical replicates and data are representative of three independent experiments. In panel f, data are means ± SD of n=6 healthy donors conducted over two independent experiments. Panels g and h are representative of two independent experiments; for gel source data, see Supplementary Figure 1. Statistical analyses were performed using unpaired, two-tailed Student’s t-tests (a-d), or a one-way ANOVA followed by a Tukey’s post-test (f). ****P ≤ 0.0001.