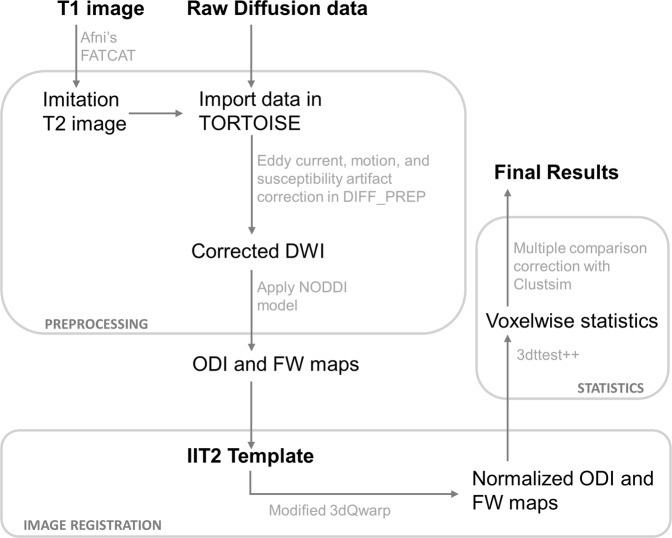
Fig. 1.
Schematic of the diffusion data processing pipeline. In preparation for DTI image registration, T1 images were processed through FATCAT in AFNI to create a volume with an approximate T2-weighted contrast. This “imitation T2” image was used solely for providing an anatomical reference. Artifacts due to bulk motion, eddy currents, and EPI susceptibility-induced geometric distortions were removed using DIFF_PREP with a single interpolation step. Prior to registration, diffusion weighted, and structural images were upsampled at a factor two and smoothed with a Perona–Malik filter to compute the transformations which were then applied to original images. Bspline correction was done using the imitation T2 image. For calculation of orientation dispersion and extracellular free water maps, we used the NODDI toolbox. For nonlinear image registration, we used a modified version of 3dQwarp in AFNI which implements an iterative refinement, where an input image is repeatedly processed through an optimizer with a final patch size of 5 mm3. AFNI′s 3dt test++ was implemented for statistical analyses, using Clustsim for multiple comparison corrections. Abbreviations: DWI diffusion weighted image, NODDI neurite orientation dispersion and density imaging, FW extracellular free water, ODI orientation dispersion index